Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | fst-RNAII/- |
| Location | 181153..181347 | Replicon | chromosome |
| Accession | NZ_CP063980 | ||
| Organism | Enterococcus faecalis strain AR_0780 | ||
Toxin (Protein)
| Gene name | fst | Uniprot ID | - |
| Locus tag | ISN59_RS00860 | Protein ID | WP_015543884.1 |
| Coordinates | 181153..181248 (+) | Length | 32 a.a. |
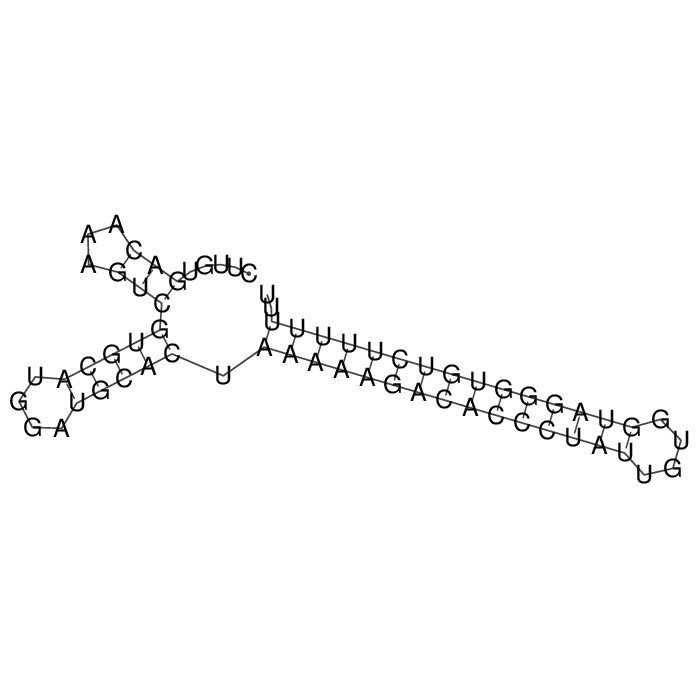
Antitoxin (RNA)
| Gene name | RNAII | ||
| Locus tag | - | ||
| Coordinates | 181283..181347 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ISN59_RS00840 (176330) | 176330..177445 | + | 1116 | WP_010709201.1 | FAD-dependent oxidoreductase | - |
| ISN59_RS00845 (177514) | 177514..178668 | - | 1155 | WP_010709200.1 | mannitol-1-phosphate 5-dehydrogenase | - |
| ISN59_RS00850 (178683) | 178683..179120 | - | 438 | WP_002355279.1 | PTS sugar transporter subunit IIA | - |
| ISN59_RS00855 (179135) | 179135..180907 | - | 1773 | WP_113843943.1 | PTS mannitol-specific transporter subunit IIBC | - |
| ISN59_RS00860 (181153) | 181153..181248 | + | 96 | WP_015543884.1 | type I toxin-antitoxin system Fst family toxin | Toxin |
| - (181283) | 181283..181347 | - | 65 | NuclAT_9 | - | Antitoxin |
| ISN59_RS00865 (181503) | 181503..181937 | - | 435 | WP_002358391.1 | PTS sugar transporter subunit IIA | - |
| ISN59_RS00870 (181948) | 181948..183981 | - | 2034 | WP_002361171.1 | PRD domain-containing protein | - |
| ISN59_RS00875 (183972) | 183972..185714 | - | 1743 | WP_162780888.1 | PTS transporter subunit EIIC | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 32 a.a. Molecular weight: 3659.47 Da Isoelectric Point: 4.6275
>T179863 WP_015543884.1 NZ_CP063980:181153-181248 [Enterococcus faecalis]
MYEIVTKILVPIFVGIVLKLVTIWLEKQNEE
MYEIVTKILVPIFVGIVLKLVTIWLEKQNEE
Download Length: 96 bp
>T179863 NZ_CP084897:c1270551-1270444 [Escherichia coli]
ATGACGCTCGCGCAGTTTGCCATGACTTTCTGGCACGACCTGGCGGCACCGATCCTGGCGGGAATTATTACCGCAGCGAT
TGTCGGCTGGTGGCGTAACCGGAAGTAA
ATGACGCTCGCGCAGTTTGCCATGACTTTCTGGCACGACCTGGCGGCACCGATCCTGGCGGGAATTATTACCGCAGCGAT
TGTCGGCTGGTGGCGTAACCGGAAGTAA
Antitoxin
Download Length: 65 bp
>AT179863 NZ_CP063980:c181347-181283 [Enterococcus faecalis]
CTTGTGACAAAGTCGTGCATGGATGCACTAAAAAGACACCCTATTGTGGTAGGGTGTCTTTTTTT
CTTGTGACAAAGTCGTGCATGGATGCACTAAAAAGACACCCTATTGTGGTAGGGTGTCTTTTTTT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|