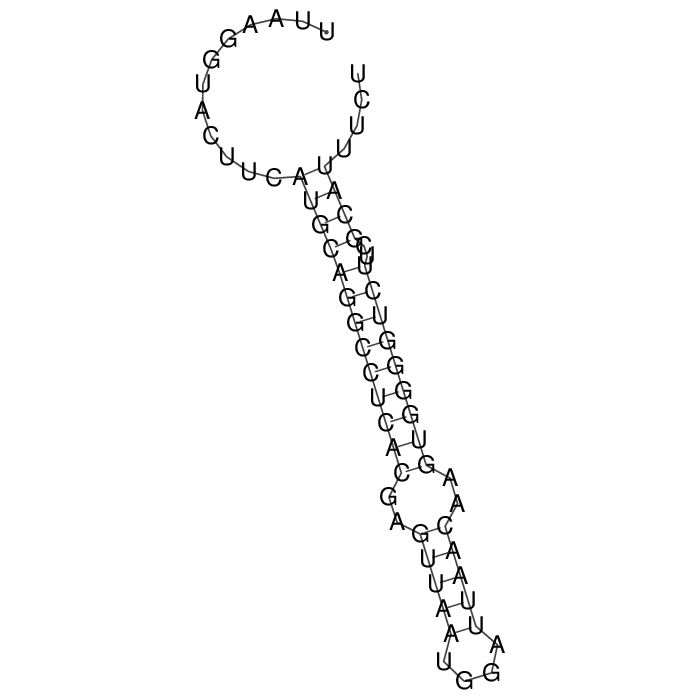
Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 78540..78794 | Replicon | plasmid unnamednovel_0 |
| Accession | NZ_CP062831 | ||
| Organism | Escherichia coli strain Res13-Lact-PER12-33-A | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | - |
| Locus tag | IMQ30_RS24490 | Protein ID | WP_032339099.1 |
| Coordinates | 78540..78746 (-) | Length | 69 a.a. |
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 78733..78794 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| IMQ30_RS24450 | 73578..74729 | - | 1152 | WP_195686650.1 | IS30-like element IS30 family transposase | - |
| IMQ30_RS24455 | 74797..75099 | - | 303 | Protein_77 | transposase | - |
| IMQ30_RS24460 | 75135..75332 | - | 198 | Protein_78 | 3'-5' exonuclease | - |
| IMQ30_RS24465 | 75372..75653 | - | 282 | WP_000969996.1 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| IMQ30_RS24470 | 75650..75919 | - | 270 | WP_000079923.1 | hypothetical protein | - |
| IMQ30_RS24475 | 76838..77694 | - | 857 | Protein_81 | incFII family plasmid replication initiator RepA | - |
| IMQ30_RS24480 | 77687..77761 | - | 75 | WP_001365705.1 | RepA leader peptide Tap | - |
| IMQ30_RS24485 | 78008..78256 | - | 249 | WP_000083838.1 | replication regulatory protein RepA | - |
| IMQ30_RS24490 | 78540..78746 | - | 207 | WP_032339099.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 78733..78794 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 78733..78794 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 78733..78794 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 78733..78794 | + | 62 | NuclAT_0 | - | Antitoxin |
| IMQ30_RS24495 | 78969..79352 | - | 384 | Protein_85 | hypothetical protein | - |
| IMQ30_RS24500 | 79470..79916 | - | 447 | WP_032339098.1 | thermonuclease family protein | - |
| IMQ30_RS24505 | 80054..81328 | - | 1275 | Protein_87 | IS66-like element ISEc23 family transposase | - |
| IMQ30_RS24515 | 82636..82980 | - | 345 | Protein_89 | IS66 family transposase | - |
| IMQ30_RS24520 | 83011..83361 | - | 351 | WP_000624722.1 | IS66 family insertion sequence element accessory protein TnpB | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Non-Mobilizable plasmid | - | - | 1..141262 | 141262 | |
| - | inside | IScluster/Tn | - | - | 72663..86490 | 13827 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 69 a.a. Molecular weight: 7808.29 Da Isoelectric Point: 8.8807
>T176767 WP_032339099.1 NZ_CP062831:c78746-78540 [Escherichia coli]
MKYLNTTDCSLFLAERSKFMTKYTLIGVLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MKYLNTTDCSLFLAERSKFMTKYTLIGVLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 207 bp
>T176767 NZ_CP082929:1979088-1979190 [Salmonella enterica subsp. enterica serovar London]
GCAAGGCGATTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
GCAAGGCGATTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
Antitoxin
Download Length: 62 bp
>AT176767 NZ_CP062831:78733-78794 [Escherichia coli]
TTAAGGTACTTCATGCAGGCCTCACGAGTTAATGGATTAACAAGTGGGGTCTTCGCATTTCT
TTAAGGTACTTCATGCAGGCCTCACGAGTTAATGGATTAACAAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|