Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 19296..19550 | Replicon | plasmid pCO_Eco4457-3 |
| Accession | NZ_CP049970 | ||
| Organism | Escherichia coli strain ARL09/232 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | - |
| Locus tag | G9448_RS25615 | Protein ID | WP_032215358.1 |
| Coordinates | 19296..19502 (-) | Length | 69 a.a. |
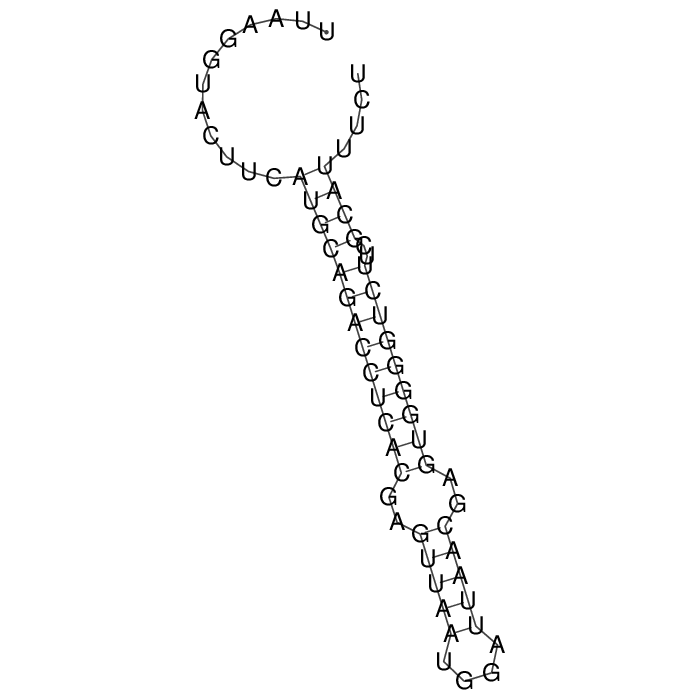
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 19489..19550 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| G9448_RS25575 | 14655..15070 | - | 416 | Protein_16 | IS1 family transposase | - |
| G9448_RS25580 | 15319..15720 | - | 402 | WP_001398199.1 | type II toxin-antitoxin system toxin endoribonuclease PemK | - |
| G9448_RS25585 | 15653..15910 | - | 258 | WP_000557619.1 | type II toxin-antitoxin system antitoxin PemI | - |
| G9448_RS25590 | 16003..16656 | - | 654 | WP_000616807.1 | CPBP family intramembrane metalloprotease | - |
| G9448_RS25595 | 16754..16894 | - | 141 | WP_001333237.1 | hypothetical protein | - |
| G9448_RS25600 | 17595..18452 | - | 858 | WP_029702152.1 | incFII family plasmid replication initiator RepA | - |
| G9448_RS25605 | 18445..18519 | - | 75 | WP_001365705.1 | RepA leader peptide Tap | - |
| G9448_RS25610 | 18755..19012 | - | 258 | WP_000083833.1 | replication regulatory protein RepA | - |
| G9448_RS25615 | 19296..19502 | - | 207 | WP_032215358.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 19489..19550 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 19489..19550 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 19489..19550 | + | 62 | NuclAT_0 | - | Antitoxin |
| - | 19489..19550 | + | 62 | NuclAT_0 | - | Antitoxin |
| G9448_RS25620 | 19689..19955 | - | 267 | WP_087757657.1 | hypothetical protein | - |
| G9448_RS25625 | 20330..20899 | - | 570 | WP_029702150.1 | DUF2726 domain-containing protein | - |
| G9448_RS25630 | 21051..21677 | - | 627 | WP_029702149.1 | hypothetical protein | - |
| G9448_RS25635 | 22089..22298 | - | 210 | WP_039023209.1 | hypothetical protein | - |
| G9448_RS25640 | 22429..22989 | - | 561 | WP_001567328.1 | fertility inhibition protein FinO | - |
| G9448_RS25645 | 23044..23790 | - | 747 | WP_052273601.1 | conjugal transfer pilus acetylase TraX | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Conjugative plasmid | blaTEM-1B | - | 1..101847 | 101847 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 69 a.a. Molecular weight: 7778.27 Da Isoelectric Point: 8.8807
>T151925 WP_032215358.1 NZ_CP049970:c19502-19296 [Escherichia coli]
MKYLNTTDCSLFLAERSKFMTKYALIGVLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
MKYLNTTDCSLFLAERSKFMTKYALIGVLAVCATVLCFSLIFRERLCELNIHRGNTVVQVTLAYEARK
Download Length: 207 bp
>T151925 NZ_CP065838:1864068-1864170 [Klebsiella quasipneumoniae]
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
Antitoxin
Download Length: 62 bp
>AT151925 NZ_CP049970:19489-19550 [Escherichia coli]
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TTAAGGTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|