Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 58600..58839 | Replicon | plasmid p124_A |
| Accession | NZ_CP048345 | ||
| Organism | Escherichia coli strain 124 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | A0A762TWR7 |
| Locus tag | GZS11_RS23055 | Protein ID | WP_023144756.1 |
| Coordinates | 58600..58734 (-) | Length | 45 a.a. |
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 58779..58839 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GZS11_RS23025 | 54313..55169 | - | 857 | Protein_66 | incFII family plasmid replication initiator RepA | - |
| GZS11_RS23035 | 55162..55236 | - | 75 | WP_031943482.1 | RepA leader peptide Tap | - |
| GZS11_RS23950 | 55233..55366 | - | 134 | Protein_68 | protein CopA/IncA | - |
| GZS11_RS23040 | 55597..57133 | + | 1537 | Protein_69 | IS21 family transposase | - |
| GZS11_RS23045 | 57148..57892 | + | 745 | Protein_70 | IS21-like element ISEc12 family helper ATPase IstB | - |
| GZS11_RS23050 | 58052..58306 | - | 255 | WP_162676766.1 | replication regulatory protein RepA | - |
| GZS11_RS23055 | 58600..58734 | - | 135 | WP_023144756.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 58779..58839 | + | 61 | NuclAT_0 | - | Antitoxin |
| - | 58779..58839 | + | 61 | NuclAT_0 | - | Antitoxin |
| - | 58779..58839 | + | 61 | NuclAT_0 | - | Antitoxin |
| - | 58779..58839 | + | 61 | NuclAT_0 | - | Antitoxin |
| GZS11_RS23060 | 58806..59092 | - | 287 | Protein_73 | DUF2726 domain-containing protein | - |
| GZS11_RS23065 | 59549..59815 | - | 267 | WP_162676767.1 | hypothetical protein | - |
| GZS11_RS23070 | 59943..60503 | - | 561 | WP_000139341.1 | fertility inhibition protein FinO | - |
| GZS11_RS23075 | 60624..62422 | - | 1799 | Protein_76 | AAA family ATPase | - |
| GZS11_RS23080 | 62392..63474 | - | 1083 | WP_060621339.1 | DUF3560 domain-containing protein | - |
| GZS11_RS23085 | 63526..63756 | - | 231 | WP_000218642.1 | hypothetical protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Non-Mobilizable plasmid | tet(B) / sul1 / qacE / aadA5 / blaTEM-1B / aac(3)-IId / mph(A) / aac(6')-Ib-cr / blaOXA-1 / blaCTX-M-15 / sul2 / aph(3'')-Ib / aph(6)-Id | - | 1..97111 | 97111 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 45 a.a. Molecular weight: 4896.90 Da Isoelectric Point: 7.7209
>T149164 WP_023144756.1 NZ_CP048345:c58734-58600 [Escherichia coli]
MTKYTLIGLLAVCATVLCFSLIFREQLCELNIHRGNTVVQVTLA
MTKYTLIGLLAVCATVLCFSLIFREQLCELNIHRGNTVVQVTLA
Download Length: 135 bp
>T149164 NZ_CP063936:c415492-415390 [Klebsiella pneumoniae]
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
GCAAGGCGACTTAGCCTGCATTAATGCCAACTTTTAGCGCACGGCTCTCTCCCAAGAGCCATTTCCCTGGACCGAATACA
GGAATCGTATTCGGTCTCTTTTT
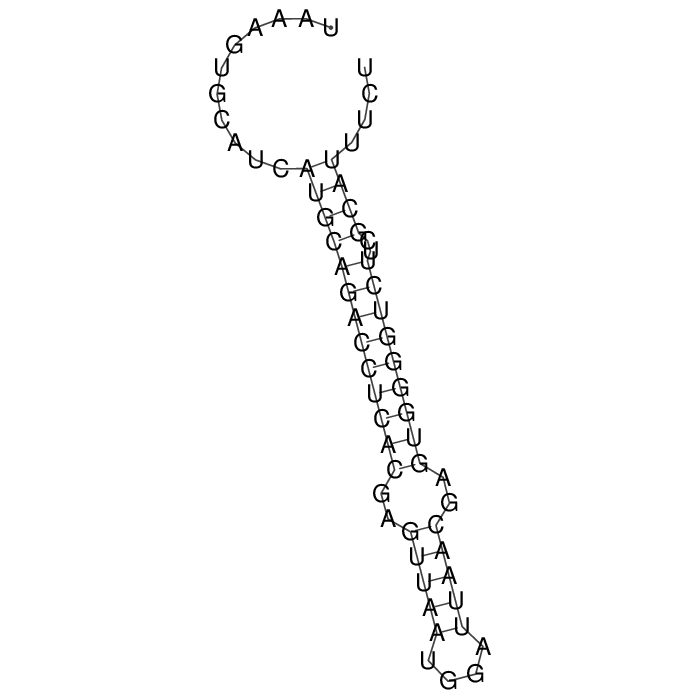
Antitoxin
Download Length: 61 bp
>AT149164 NZ_CP048345:58779-58839 [Escherichia coli]
TAAAGTGCATCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
TAAAGTGCATCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|