Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | symER/SymE(toxin) |
| Location | 4913651..4914063 | Replicon | chromosome |
| Accession | NZ_CP038372 | ||
| Organism | Escherichia coli O157:H7 strain F6294 | ||
Toxin (Protein)
| Gene name | symE | Uniprot ID | Q8FA88 |
| Locus tag | E3161_RS24805 | Protein ID | WP_000132630.1 |
| Coordinates | 4913722..4914063 (+) | Length | 114 a.a. |
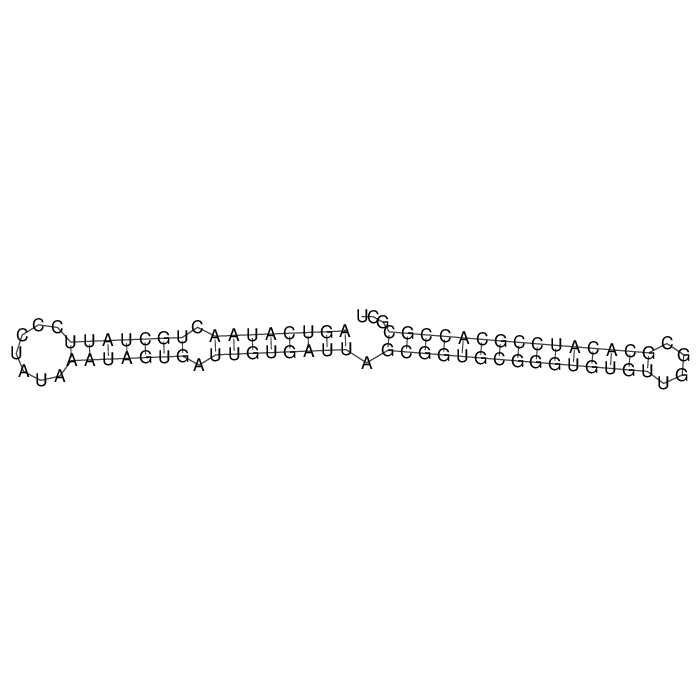
Antitoxin (RNA)
| Gene name | symR | ||
| Locus tag | - | ||
| Coordinates | 4913651..4913727 (-) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| E3161_RS24795 | 4910278..4911747 | + | 1470 | WP_001302468.1 | type I restriction-modification system subunit M | - |
| E3161_RS24800 | 4911747..4913501 | + | 1755 | WP_000110082.1 | restriction endonuclease subunit S | - |
| - | 4913651..4913727 | - | 77 | NuclAT_16 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_16 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_16 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_16 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_17 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_17 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_17 | - | Antitoxin |
| - | 4913651..4913727 | - | 77 | NuclAT_17 | - | Antitoxin |
| E3161_RS24805 | 4913722..4914063 | + | 342 | WP_000132630.1 | endoribonuclease SymE | Toxin |
| E3161_RS24810 | 4914110..4915273 | - | 1164 | WP_001304006.1 | DUF1524 domain-containing protein | - |
| E3161_RS24815 | 4915321..4916202 | - | 882 | WP_001304007.1 | DUF262 domain-containing protein | - |
| E3161_RS24820 | 4916299..4916463 | - | 165 | WP_000394274.1 | DUF1127 domain-containing protein | - |
| E3161_RS24825 | 4916640..4918052 | + | 1413 | WP_000199295.1 | PLP-dependent aminotransferase family protein | - |
| E3161_RS24830 | 4918295..4918495 | - | 201 | WP_001310455.1 | hypothetical protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| inside | Genomic island | - | espX6 | 4905850..4927089 | 21239 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 114 a.a. Molecular weight: 12294.13 Da Isoelectric Point: 8.4982
>T122106 WP_000132630.1 NZ_CP038372:4913722-4914063 [Escherichia coli O157:H7]
MTDTHSIAQPFEAEVSPANNRQLTVSYASRYPDYSRIPAITLKGQWLEAAGFATGTVVDVKVMEGCIVLTAQPPAAAESE
LMQSLRQVCKLSARKQRQVQEFIGVIAGKQKVA
MTDTHSIAQPFEAEVSPANNRQLTVSYASRYPDYSRIPAITLKGQWLEAAGFATGTVVDVKVMEGCIVLTAQPPAAAESE
LMQSLRQVCKLSARKQRQVQEFIGVIAGKQKVA
Download Length: 342 bp
>T122106 NZ_CP038372:4913722-4914063 [Escherichia coli O157:H7]
ATGACTGATACGCATTCTATTGCACAACCGTTCGAAGCAGAAGTCTCCCCGGCAAATAACCGTCAATTAACCGTGAGTTA
TGCGAGTCGTTACCCGGATTACAGCCGTATTCCCGCCATCACCCTGAAAGGTCAGTGGCTGGAAGCCGCCGGTTTTGCCA
CCGGCACGGTGGTAGATGTCAAAGTGATGGAAGGCTGTATTGTCCTCACCGCCCAACCACCCGCCGCCGCCGAGAGCGAA
CTGATGCAGTCGCTGCGCCAGGTGTGCAAGCTGTCGGCGCGTAAACAAAGGCAGGTGCAGGAGTTTATTGGGGTGATTGC
GGGTAAACAGAAAGTTGCCTGA
ATGACTGATACGCATTCTATTGCACAACCGTTCGAAGCAGAAGTCTCCCCGGCAAATAACCGTCAATTAACCGTGAGTTA
TGCGAGTCGTTACCCGGATTACAGCCGTATTCCCGCCATCACCCTGAAAGGTCAGTGGCTGGAAGCCGCCGGTTTTGCCA
CCGGCACGGTGGTAGATGTCAAAGTGATGGAAGGCTGTATTGTCCTCACCGCCCAACCACCCGCCGCCGCCGAGAGCGAA
CTGATGCAGTCGCTGCGCCAGGTGTGCAAGCTGTCGGCGCGTAAACAAAGGCAGGTGCAGGAGTTTATTGGGGTGATTGC
GGGTAAACAGAAAGTTGCCTGA
Antitoxin
Download Length: 77 bp
>AT122106 NZ_CP038372:c4913727-4913651 [Escherichia coli O157:H7]
AGTCATAACTGCTATTCCCTATAAATAGTGATTGTGATTAGCGGTGCGGGTGTGTTGGCGCACATCCGCACCGCGCT
AGTCATAACTGCTATTCCCTATAAATAGTGATTGTGATTAGCGGTGCGGGTGTGTTGGCGCACATCCGCACCGCGCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|