Detailed information of TA system
Overview
TA module
| Type | I | Classification (family/domain) | hok-sok/- |
| Location | 39228..39482 | Replicon | plasmid unnamed1 |
| Accession | NZ_CP030779 | ||
| Organism | Escherichia albertii strain 05-3106 | ||
Toxin (Protein)
| Gene name | srnB | Uniprot ID | - |
| Locus tag | CYM74_RS23475 | Protein ID | WP_000876345.1 |
| Coordinates | 39228..39434 (-) | Length | 69 a.a. |
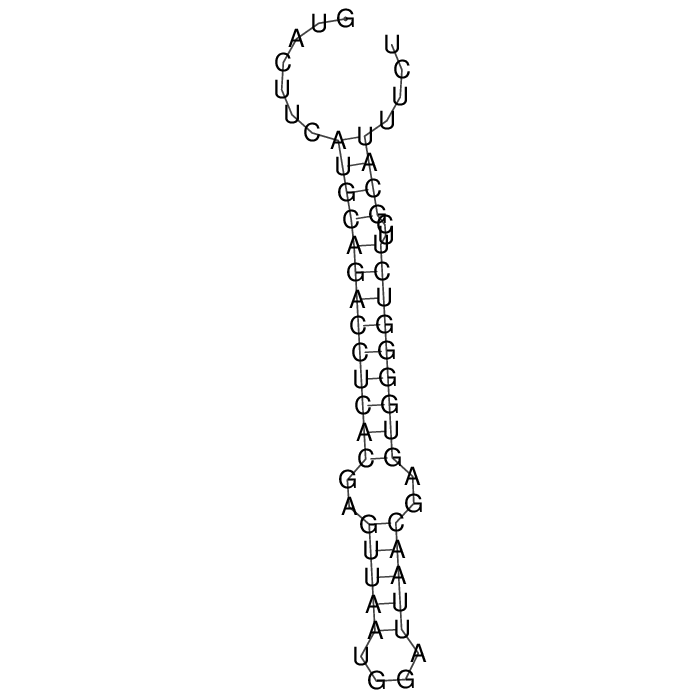
Antitoxin (RNA)
| Gene name | srnC | ||
| Locus tag | - | ||
| Coordinates | 39426..39482 (+) |
Genomic Context
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CYM74_RS23430 | 34603..35085 | - | 483 | WP_001311056.1 | hypothetical protein | - |
| CYM74_RS23435 | 35202..36013 | - | 812 | Protein_39 | 3'-5' exoribonuclease | - |
| CYM74_RS23440 | 36059..36340 | - | 282 | WP_044718080.1 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| CYM74_RS23445 | 36337..36606 | - | 270 | WP_000079928.1 | hypothetical protein | - |
| CYM74_RS23455 | 37522..38379 | - | 858 | WP_000130987.1 | incFII family plasmid replication initiator RepA | - |
| CYM74_RS23460 | 38372..38446 | - | 75 | WP_001365705.1 | RepA leader peptide Tap | - |
| CYM74_RS23470 | 38684..38944 | - | 261 | WP_000078108.1 | replication regulatory protein RepA | - |
| CYM74_RS23475 | 39228..39434 | - | 207 | WP_000876345.1 | type I toxin-antitoxin system Hok family toxin | Toxin |
| - | 39426..39482 | + | 57 | NuclAT_0 | - | Antitoxin |
| - | 39426..39482 | + | 57 | NuclAT_0 | - | Antitoxin |
| - | 39426..39482 | + | 57 | NuclAT_0 | - | Antitoxin |
| - | 39426..39482 | + | 57 | NuclAT_0 | - | Antitoxin |
| CYM74_RS23480 | 39723..39932 | - | 210 | WP_024234785.1 | hemolysin expression modulator Hha | - |
| CYM74_RS23485 | 39988..40224 | - | 237 | WP_000376558.1 | hypothetical protein | - |
| CYM74_RS23490 | 40311..41324 | - | 1014 | WP_000118342.1 | hypothetical protein | - |
| CYM74_RS23495 | 41431..41697 | - | 267 | WP_113623322.1 | hypothetical protein | - |
| CYM74_RS23500 | 41736..42518 | - | 783 | WP_001317493.1 | IS21-like element IS100kyp family helper ATPase IstB | - |
| CYM74_RS23505 | 42515..43537 | - | 1023 | WP_000255946.1 | IS21-like element IS100 family transposase | - |
| CYM74_RS23510 | 43604..43777 | - | 174 | WP_206430860.1 | hypothetical protein | - |
| CYM74_RS23520 | 44171..44419 | + | 249 | WP_000618110.1 | DinI-like family protein | - |
Associated MGEs
| MGE detail |
Similar MGEs |
Relative position |
MGE Type | Cargo ARG | Virulence gene | Coordinates | Length (bp) |
|---|---|---|---|---|---|---|---|
| - | inside | Non-Mobilizable plasmid | - | bfpF / bfpE / bfpD / bfpA | 1..56603 | 56603 |
Relative position:
(1) inside: TA loci is completely located inside the MGE;
(2) overlap: TA loci is partially overlapped with the MGE;
(3) flank: The TA loci is located in the 5 kb flanking regions of MGE.
Sequences
Toxin
Download Length: 69 a.a. Molecular weight: 7806.32 Da Isoelectric Point: 8.8807
>T107969 WP_000876345.1 NZ_CP030779:c39434-39228 [Escherichia albertii]
MKYLNTTDCSLFLAERSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQITLAYEARK
MKYLNTTDCSLFLAERSKFMTKYALIGLLAVCATVLCFSLIFRERLCELNIHRGNTVVQITLAYEARK
Download Length: 207 bp
>T107969 NZ_CP030779:c39434-39228 [Escherichia albertii]
ATGAAGTACTTGAACACTACTGATTGTAGCCTCTTCCTTGCAGAGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTGTGTGAGCTGAATATTCACAGGG
GAAATACAGTGGTGCAGATAACTCTGGCCTACGAAGCACGGAAGTAA
ATGAAGTACTTGAACACTACTGATTGTAGCCTCTTCCTTGCAGAGAGGTCAAAGTTTATGACGAAATATGCCCTTATCGG
GTTGCTCGCCGTGTGCGCCACGGTGTTGTGTTTTTCACTGATATTCAGGGAACGGTTGTGTGAGCTGAATATTCACAGGG
GAAATACAGTGGTGCAGATAACTCTGGCCTACGAAGCACGGAAGTAA
Antitoxin
Download Length: 57 bp
>AT107969 NZ_CP030779:39426-39482 [Escherichia albertii]
GTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
GTACTTCATGCAGACCTCACGAGTTAATGGATTAACGAGTGGGGTCTTCGCATTTCT
Similar Proteins
Only experimentally validated proteins are listed.
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|
| Protein | Organism | Identities (%) | Coverage (%) | Ha-value |
|---|