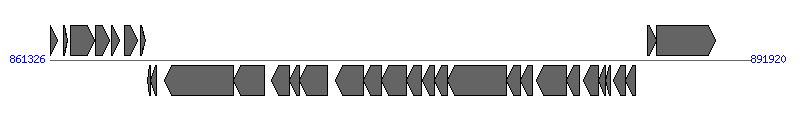| Component | TraA_F | TraB_F | TraC_F | TraD_F | TraE_F | TraF_F | TraG_F | TraH_F | TraI_F |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| Component | TraK_F | TraL_F | TraM_F | TraN_F | TraQ_F | TraU_F | TraV_F | TraW_F | TraX_F |
| Number | 1 | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 0 |
| Component | TraY_F | TrbC_F | Orf169_F |
| Number | 0 | 1 | 1 |
The information of T4SS components from NC_011035 |
 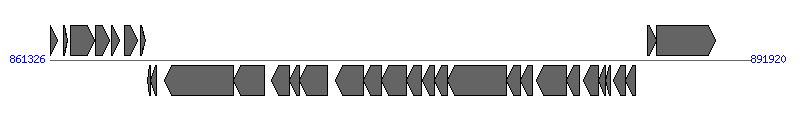 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | NGK_1058 | 861328..861639 [+], 312 | 194098621 | hypothetical protein | | |
| 2 | NGK_1059 | 861911..862090 [+], 180 | 194098622 | hypothetical protein | | |
| 3 | NGK_1060 | 862205..863275 [+], 1071 | 194098623 | hypothetical protein | | |
| 4 | NGK_1061 | 863328..863942 [+], 615 | 194098624 | hypothetical protein | | |
| 5 | NGK_1062 | 863997..864359 [+], 363 | 194098625 | hypothetical protein | | |
| 6 | NGK_1063 | 864563..865150 [+], 588 | 194098626 | CspA protein | | |
| 7 | NGK_1064 | 865275..865520 [+], 246 | 194098627 | hypothetical protein | | |
| 8 | NGK_1065 | 865577..865735 [-], 159 | 194098628 | hypothetical protein | | |
| 9 | NGK_1066 | 865739..865996 [-], 258 | 194098629 | AtlA protein | | |
| 10 | NGK_1067 | 866326..869328 [-], 3003 | 194098630 | protein TraG | TraG_F | |
| 11 | NGK_1068 | 869335..870693 [-], 1359 | 194098631 | protein TraH | TraH_F | |
| 12 | NGK_1069 | 871018..871794 [-], 777 | 194098632 | protein TraF | TraF_F | |
| 13 | NGK_1070 | 871806..872231 [-], 426 | 194098633 | hypothetical protein | | |
| 14 | NGK_1071 | 872218..873471 [-], 1254 | 194098634 | protein TraN | TraN_F | |
| 15 | NGK_1072 | 873798..875042 [-], 1245 | 194098635 | hypothetical protein | | |
| 16 | NGK_1073 | 875039..875794 [-], 756 | 194098636 | protein TrbC | TrbC_F | |
| 17 | NGK_1074 | 875806..876924 [-], 1119 | 194098637 | protein TraU | TraU_F | |
| 18 | NGK_1075 | 876921..877577 [-], 657 | 194098638 | protein TraW | TraW_F | |
| 19 | NGK_1076 | 877586..878131 [-], 546 | 194098639 | protein TrbI | | |
| 20 | NGK_1077 | 878131..878679 [-], 549 | 194098640 | hypothetical protein | | |
| 21 | NGK_1078 | 878707..881283 [-], 2577 | 194098641 | protein TraC | TraC_F | |
| 22 | NGK_1079 | 881293..881898 [-], 606 | 194098642 | protein TraV | TraV_F | |
| 23 | NGK_1080 | 881871..882431 [-], 561 | 194098643 | protein DsbC | | |
| 24 | NGK_1081 | 882600..883913 [-], 1314 | 194098644 | protein TraB | TraB_F | |
| 25 | NGK_1082 | 883915..884472 [-], 558 | 194098645 | protein TraK | TraK_F | |
| 26 | NGK_1083 | 884651..885301 [-], 651 | 194098646 | protein TraE | TraE_F | |
| 27 | NGK_1084 | 885313..885594 [-], 282 | 194098647 | protein TraL | TraL_F | |
| 28 | NGK_1085 | 885616..885840 [-], 225 | 194098648 | TraA protein | TraA_F | |
| 29 | NGK_1086 | 885968..886489 [-], 522 | 194098649 | hypothetical protein | | |
| 30 | NGK_1087 | 886459..886920 [-], 462 | 194098650 | protein LtgX | Orf169_F | |
| 31 | NGK_1088 | 887431..887850 [+], 420 | 194098651 | hypothetical protein | | |
| 32 | NGK_1089 | 887847..890399 [+], 2553 | 194098652 | TraI | | |
| |
 This T4SS contains information of accessory protein. This T4SS contains information of accessory protein. |
 Genes in the 5-Kb flanking regions if available, or non-essential genes in the T4SS gene cluster if any. Genes in the 5-Kb flanking regions if available, or non-essential genes in the T4SS gene cluster if any. |