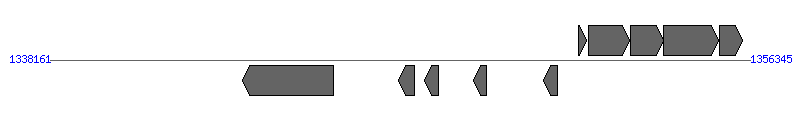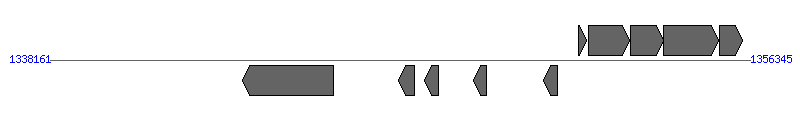The information of T4SS components from NC_006831 |
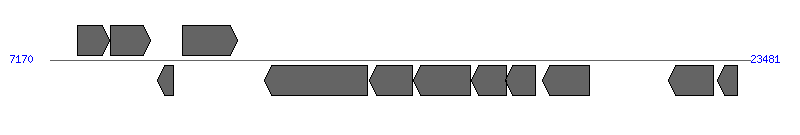
| Region 1: 12170..18481 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERGA_CDS_00090 | 7804..8553 [+], 750 | 58616736 | hypothetical protein | | |
| 2 | ERGA_CDS_00100 (nadA) | 8568..9515 [+], 948 | 58616737 | quinolinate synthetase | | |
| 3 | ERGA_CDS_00110 (fdxA) | 9675..10052 [-], 378 | 58616738 | ferredoxin | | |
| 4 | ERGA_CDS_00120 | 10263..11528 [+], 1266 | 58616739 | hypothetical protein | | |
| 5 | ERGA_CDS_00130 (virD4) | 12170..14572 [-], 2403 | 58616740 | type IV secretion system component VirD4 | VirD4 | |
| 6 | ERGA_CDS_00140 (virB11) | 14613..15611 [-], 999 | 58616741 | type IV secretion system ATPase VirB11 | VirB11 | |
| 7 | ERGA_CDS_00150 (virB10) | 15635..16975 [-], 1341 | 58616742 | VirB10 protein | VirB10 | |
| 8 | ERGA_CDS_00160 (trbG) | 16996..17799 [-], 804 | 58616743 | conjugal transfer protein trbG precursor | VirB9 | |
| 9 | ERGA_CDS_00170 | 17783..18481 [-], 699 | 58616744 | hypothetical protein | VirB8 | |
| 10 | ERGA_CDS_00180 (ribA) | 18637..19752 [-], 1116 | 58616745 | GTP cyclohydrolase II | | |
| 11 | ERGA_CDS_00190 | 21573..22637 [-], 1065 | 58616746 | hypothetical protein | | |
| 12 | ERGA_CDS_00200 | 22713..23192 [-], 480 | 58616747 | hypothetical protein | | |
| |
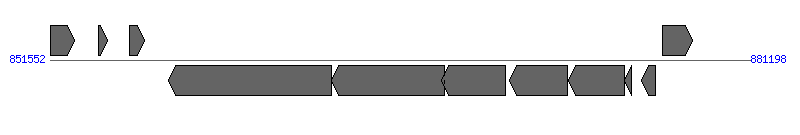
| Region 2: 856552..876198 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERGA_CDS_05330 (ispH) | 851584..852582 [+], 999 | 58617260 | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | | |
| 2 | ERGA_CDS_05340 (dut) | 853598..853972 [+], 375 | 58617261 | deoxyuridine 5-triphosphate nucleotidohydrolase | | |
| 3 | ERGA_CDS_05350 | 854936..855535 [+], 600 | 58617262 | hypothetical protein | | |
| 4 | ERGA_CDS_05360 | 856552..863454 [-], 6903 | 58617263 | hypothetical protein | VirB6 | |
| 5 | ERGA_CDS_05370 | 863472..868247 [-], 4776 | 58617264 | hypothetical protein | VirB6 | |
| 6 | ERGA_CDS_05380 | 868129..870861 [-], 2733 | 58617265 | hypothetical protein | VirB6 | |
| 7 | ERGA_CDS_05390 | 870999..873464 [-], 2466 | 58617266 | hypothetical protein | VirB6 | |
| 8 | ERGA_CDS_05400 (virB4) | 873497..875902 [-], 2406 | 58617267 | type IV secretion system ATPase VirB4 | VirB4 | |
| 9 | ERGA_CDS_05410 | 875899..876198 [-], 300 | 58617268 | type IV secretion system protein VirB3 | VirB3 | |
| 10 | ERGA_CDS_05420 (sodB) | 876584..877216 [-], 633 | 58617269 | superoxide dismutase | | |
| 11 | ERGA_CDS_05430 (pstA) | 877482..878744 [+], 1263 | 58617270 | phosphate ABC transporter permease | | |
| |
| Region 3: 1254693..1255469 |
 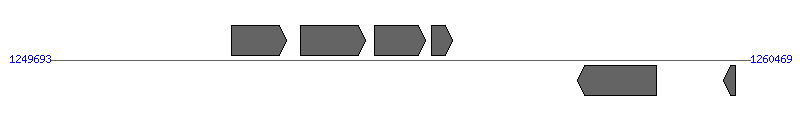 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERGA_CDS_07820 | 1252488..1253333 [+], 846 | 58617509 | hypothetical protein | | |
| 2 | ERGA_CDS_07830 (pdhA) | 1253556..1254545 [+], 990 | 58617510 | pyruvate dehydrogenase E1 component, alpha subunit | | |
| 3 | ERGA_CDS_07840 (trbG) | 1254693..1255469 [+], 777 | 58617511 | conjugal transfer protein TRBG precursor | VirB9 | |
| 4 | ERGA_CDS_07850 (trx) | 1255560..1255883 [+], 324 | 58617512 | thioredoxin | | |
| 5 | ERGA_CDS_07860 | 1257808..1259025 [-], 1218 | 58617513 | hypothetical protein | | |
| 6 | ERGA_CDS_07870 (xseB) | 1260059..1260247 [-], 189 | 58617514 | exodeoxyribonuclease VII small subunit | | |
| |
| Region 4: 1343161..1351345 |
 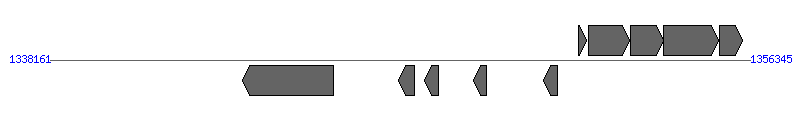 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERGA_CDS_08350 (virB4) | 1343161..1345533 [-], 2373 | 58617562 | VirB4 protein | VirB4 | |
| 2 | ERGA_CDS_08360 | 1347206..1347622 [-], 417 | 58617563 | hypothetical protein | VirB2 | |
| 3 | ERGA_CDS_08370 | 1347876..1348241 [-], 366 | 58617564 | hypothetical protein | VirB2 | |
| 4 | ERGA_CDS_08380 | 1349154..1349495 [-], 342 | 58617565 | hypothetical protein | VirB2 | |
| 5 | ERGA_CDS_08390 | 1350971..1351345 [-], 375 | 58617566 | hypothetical protein | VirB2 | |
| 6 | ERGA_CDS_08400 | 1351895..1352095 [+], 201 | 58617567 | hypothetical protein | | |
| 7 | ERGA_CDS_08410 (hflK) | 1352150..1353220 [+], 1071 | 58617568 | protease activity modulator hflk | | |
| 8 | ERGA_CDS_08420 (hflC) | 1353232..1354104 [+], 873 | 58617569 | Hflc protein | | |
| 9 | ERGA_CDS_08430 (degP) | 1354104..1355534 [+], 1431 | 58617570 | serine protease do-like precursor | | |
| 10 | ERGA_CDS_08440 | 1355542..1356156 [+], 615 | 58617571 | hypothetical protein | | |
| |
| Region 5: 1343161..1351345 |
 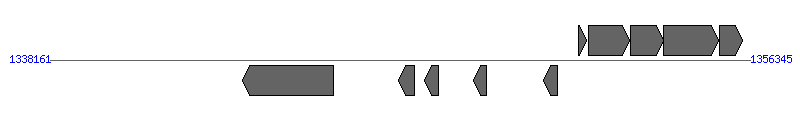 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERGA_CDS_08350 (virB4) | 1343161..1345533 [-], 2373 | 58617562 | VirB4 protein | VirB4 | |
| 2 | ERGA_CDS_08360 | 1347206..1347622 [-], 417 | 58617563 | hypothetical protein | VirB2 | |
| 3 | ERGA_CDS_08370 | 1347876..1348241 [-], 366 | 58617564 | hypothetical protein | VirB2 | |
| 4 | ERGA_CDS_08380 | 1349154..1349495 [-], 342 | 58617565 | hypothetical protein | VirB2 | |
| 5 | ERGA_CDS_08390 | 1350971..1351345 [-], 375 | 58617566 | hypothetical protein | VirB2 | |
| 6 | ERGA_CDS_08400 | 1351895..1352095 [+], 201 | 58617567 | hypothetical protein | | |
| 7 | ERGA_CDS_08410 (hflK) | 1352150..1353220 [+], 1071 | 58617568 | protease activity modulator hflk | | |
| 8 | ERGA_CDS_08420 (hflC) | 1353232..1354104 [+], 873 | 58617569 | Hflc protein | | |
| 9 | ERGA_CDS_08430 (degP) | 1354104..1355534 [+], 1431 | 58617570 | serine protease do-like precursor | | |
| 10 | ERGA_CDS_08440 | 1355542..1356156 [+], 615 | 58617571 | hypothetical protein | | |
| |