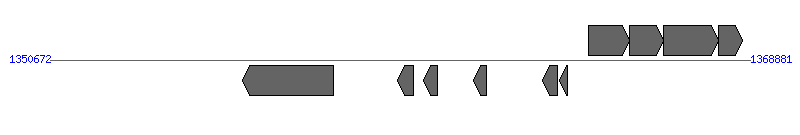The information of T4SS components from NC_006832 |
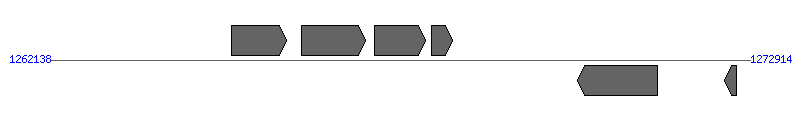
| Region 1: 12114..18434 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERWE_CDS_00090 | 7828..8577 [+], 750 | 58578673 | hypothetical protein | | |
| 2 | ERWE_CDS_00100 (nadA) | 8592..9539 [+], 948 | 58578674 | quinolinate synthetase | | |
| 3 | ERWE_CDS_00110 (fdxA) | 9699..10076 [-], 378 | 58578675 | ferredoxin | | |
| 4 | ERWE_CDS_00120 | 10287..11462 [+], 1176 | 58578676 | hypothetical protein | | |
| 5 | ERWE_CDS_00130 (virD4) | 12114..14519 [-], 2406 | 58578677 | type IV secretion system component VirD4 | VirD4 | |
| 6 | ERWE_CDS_00140 (virB11) | 14560..15558 [-], 999 | 58578678 | type IV secretion system ATPase VirB11 | VirB11 | |
| 7 | ERWE_CDS_00150 (virB10) | 15582..16928 [-], 1347 | 58578679 | VirB10 protein | VirB10 | |
| 8 | ERWE_CDS_00160 (trbG) | 16949..17752 [-], 804 | 58578680 | conjugal transfer protein trbG precursor | VirB9 | |
| 9 | ERWE_CDS_00170 | 17736..18434 [-], 699 | 58578681 | hypothetical protein | VirB8 | |
| 10 | ERWE_CDS_00180 (ribA) | 18590..19705 [-], 1116 | 58578682 | GTP cyclohydrolase II | | |
| 11 | ERWE_CDS_00190 | 21492..22556 [-], 1065 | 58578683 | hypothetical protein | | |
| 12 | ERWE_CDS_00200 | 22632..23111 [-], 480 | 58578684 | hypothetical protein | | |
| |
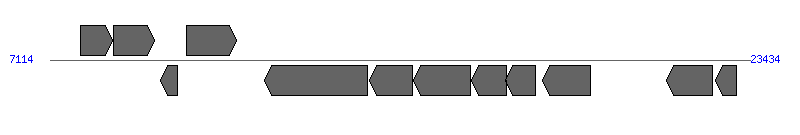
| Region 2: 863430..880589 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERWE_CDS_05450 | 859350..859961 [+], 612 | 58579209 | hypothetical protein | | |
| 2 | ERWE_CDS_05460 | 860977..863337 [-], 2361 | 58579210 | hypothetical protein | | |
| 3 | ERWE_CDS_05470 | 863430..867902 [-], 4473 | 58579211 | hypothetical protein | VirB6 | |
| 4 | ERWE_CDS_05480 | 867920..872629 [-], 4710 | 58579212 | hypothetical protein | VirB6 | |
| 5 | ERWE_CDS_05490 | 872511..875246 [-], 2736 | 58579213 | hypothetical protein | VirB6 | |
| 6 | ERWE_CDS_05500 | 875384..877855 [-], 2472 | 58579214 | hypothetical protein | VirB6 | |
| 7 | ERWE_CDS_05510 (virB4) | 877888..880293 [-], 2406 | 58579215 | type IV secretion system ATPase VirB4 | VirB4 | |
| 8 | ERWE_CDS_05520 | 880290..880589 [-], 300 | 58579216 | type IV secretion system protein VirB3 | VirB3 | |
| 9 | ERWE_CDS_05530 (sodB) | 880972..881616 [-], 645 | 58579217 | superoxide dismutase | | |
| 10 | ERWE_CDS_05540 (pstA) | 881869..883131 [+], 1263 | 58579218 | phosphate ABC transporter permease | | |
| |
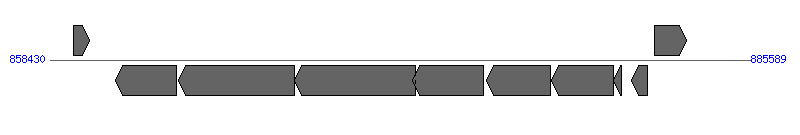
| Region 3: 1267138..1267914 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERWE_CDS_07910 | 1264937..1265782 [+], 846 | 58579455 | hypothetical protein | | |
| 2 | ERWE_CDS_07920 (pdhA) | 1266004..1266990 [+], 987 | 58579456 | pyruvate dehydrogenase E1 component, alpha subunit | | |
| 3 | ERWE_CDS_07930 (trbG) | 1267138..1267914 [+], 777 | 58579457 | conjugal transfer protein TRBG precursor | VirB9 | |
| 4 | ERWE_CDS_07940 (trx) | 1268005..1268328 [+], 324 | 58579458 | thioredoxin | | |
| 5 | ERWE_CDS_07950 | 1270253..1271494 [-], 1242 | 58579459 | hypothetical protein | | |
| 6 | ERWE_CDS_07960 (xseB) | 1272523..1272711 [-], 189 | 58579460 | exodeoxyribonuclease VII small subunit | | |
| |
| Region 4: 1355672..1363881 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | ERWE_CDS_08440 (virB4) | 1355672..1358044 [-], 2373 | 58579508 | VirB4 protein | VirB4 | |
| 2 | ERWE_CDS_08450 | 1359722..1360132 [-], 411 | 58579509 | hypothetical protein | VirB2 | |
| 3 | ERWE_CDS_08460 | 1360393..1360758 [-], 366 | 58579510 | hypothetical protein | VirB2 | |
| 4 | ERWE_CDS_08470 | 1361676..1362038 [-], 363 | 58579511 | hypothetical protein | VirB2 | |
| 5 | ERWE_CDS_08480 | 1363495..1363881 [-], 387 | 58579512 | hypothetical protein | VirB2 | |
| 6 | ERWE_CDS_08490 | 1363936..1364145 [-], 210 | 58579513 | hypothetical protein | | |
| 7 | ERWE_CDS_08500 (hflK) | 1364675..1365745 [+], 1071 | 58579514 | protease activity modulator hflk | | |
| 8 | ERWE_CDS_08510 (hflC) | 1365757..1366629 [+], 873 | 58579515 | Hflc protein | | |
| 9 | ERWE_CDS_08520 (degP) | 1366629..1368059 [+], 1431 | 58579516 | serine protease do-like precursor | | |
| 10 | ERWE_CDS_08530 | 1368067..1368681 [+], 615 | 58579517 | hypothetical protein | | |
| |