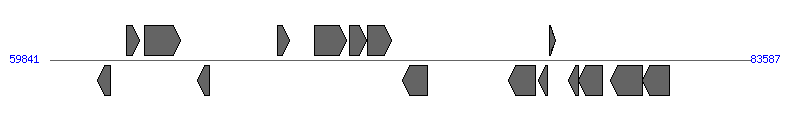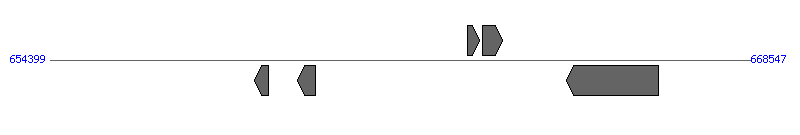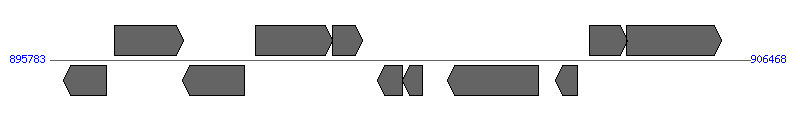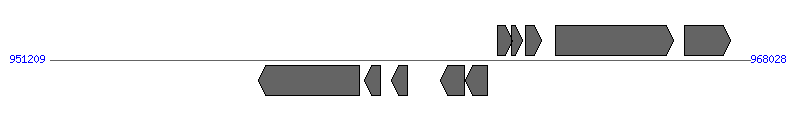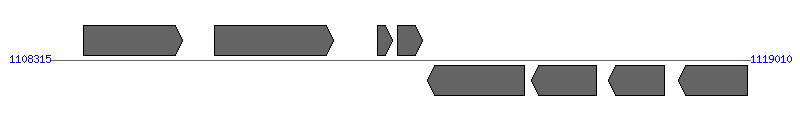The information of T4SS components from NC_004842 |
| Region 1: 23496..48535 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM026 | 21386..22900 [+], 1515 | 56416391 | monovalent cation/H+ antiporter subunit D | | |
| 2 | AM027 | 22915..23301 [-], 387 | 56416392 | hypothetical protein | | |
| 3 | AM030 (orfX) | 23496..23882 [-], 387 | 56416393 | hypothetical protein | VirB2 | |
| 4 | AM032 (orfY) | 23955..24188 [-], 234 | 56416394 | hypothetical protein | VirB2 | |
| 5 | AM034 | 25004..27445 [-], 2442 | 56416395 | hypothetical protein | | |
| 6 | AM035 | 27643..27852 [+], 210 | 56416396 | hypothetical protein | | |
| 7 | AM037 (thiG) | 27915..28709 [+], 795 | 56416397 | thiazole synthase | | |
| 8 | AM038 | 28807..29247 [-], 441 | 56416398 | hypothetical protein | | |
| 9 | AM039 (gltX) | 29429..30754 [+], 1326 | 56416399 | glutamyl-tRNA synthetase | | |
| 10 | AM041 | 31186..32391 [+], 1206 | 56416400 | hypothetical protein | | |
| 11 | AM042 | 32410..32967 [+], 558 | 56416401 | hypothetical protein | | |
| 12 | AM044 (orfX) | 33143..33541 [-], 399 | 56416402 | hypothetical protein | VirB2 | |
| 13 | AM046 (orfY) | 33614..33847 [-], 234 | 56416403 | hypothetical protein | VirB2 | |
| 14 | AM051 (rho) | 36761..38125 [-], 1365 | 56416404 | transcription termination factor Rho | | |
| 15 | AM052 | 38234..38851 [-], 618 | 56416405 | hypothetical protein | | |
| 16 | AM053 (rpmI) | 39269..39469 [+], 201 | 56416406 | 50S ribosomal protein L35 | | |
| 17 | AM054 (rplT) | 39496..39882 [+], 387 | 56416407 | 50S ribosomal protein L20 | | |
| 18 | AM055 (pheS) | 39845..40900 [+], 1056 | 56416408 | phenylalanyl-tRNA synthetase subunit alpha | | |
| 19 | AM056 | 40991..41749 [+], 759 | 56416409 | hypothetical protein | | |
| 20 | AM058 (glmU) | 41930..43216 [+], 1287 | 56416410 | UDP-N-acetylglucosamine pyrophosphorylase | | |
| 21 | AM059 (rpsP) | 43369..43638 [-], 270 | 56416411 | 30S ribosomal protein S16 | | |
| 22 | AM060 (proP1) | 43929..45215 [+], 1287 | 56416412 | proline/betaine transport protein | | |
| 23 | AM063 (orfY) | 47834..48067 [+], 234 | 56416413 | hypothetical protein | VirB2 | |
| 24 | AM065 (orfX) | 48140..48535 [+], 396 | 56416414 | hypothetical protein | VirB2 | |
| 25 | AM069 (fpbA) | 49429..50469 [+], 1041 | 56416415 | iron binding protein FbpA precursor | | |
| 26 | AM070 | 50470..50649 [+], 180 | 56416416 | hypothetical protein | | |
| |
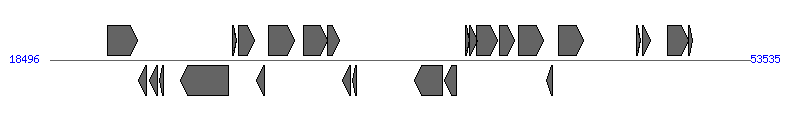
| Region 2: 64841..78587 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM073 (nrdR) | 61443..61898 [-], 456 | 56416419 | NrdR family transcriptional regulator | | |
| 2 | AM074 | 62431..62886 [+], 456 | 56416420 | hypothetical protein | | |
| 3 | AM075 (omp14) | 63055..64269 [+], 1215 | 56416421 | outer membrane protein 14 | | |
| 4 | AM077 (orfX) | 64841..65242 [-], 402 | 56416422 | hypothetical protein | VirB2 | |
| 5 | AM082 (orfX) | 67571..67978 [+], 408 | 56416423 | hypothetical protein | VirB2 | |
| 6 | AM085 (recA) | 68810..69889 [+], 1080 | 56416424 | recombinase A | | |
| 7 | AM087 (pyrE) | 69984..70586 [+], 603 | 56416425 | orotate phosphoribosyltransferase | | |
| 8 | AM088 (thiF) | 70620..71426 [+], 807 | 56416426 | thiamine biosynthesis protein | | |
| 9 | AM090 (msp4) | 71782..72630 [-], 849 | 56416427 | major surface protein 4 | | |
| 10 | AM092 | 75388..76326 [-], 939 | 56416428 | hypothetical protein | | |
| 11 | AM093 | 76411..76731 [-], 321 | 56416429 | hypothetical protein | | |
| 12 | AM094 | 76781..76984 [+], 204 | 56416430 | hypothetical protein | | |
| 13 | AM096 (trxA) | 77419..77766 [-], 348 | 56416431 | thioredoxin | | |
| 14 | AM097 (trbG) | 77775..78587 [-], 813 | 56416432 | conjugal transfer protein trbG precursor | VirB9 | |
| 15 | AM101 (pdhA) | 78838..79956 [-], 1119 | 56416433 | pyruvate dehydrogenase E1 component, subunit alpha precursor | | |
| 16 | AM102 (rrf) | 79928..80869 [-], 942 | 56416434 | hypothetical protein | | |
| |
| Region 3: 175825..176214 |
 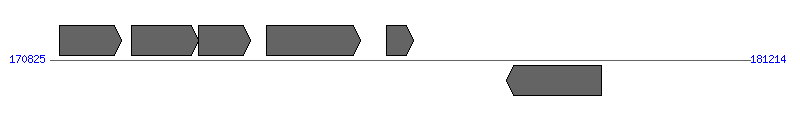 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM203 (gshB) | 170965..171891 [+], 927 | 56416507 | glutathione synthetase | | |
| 2 | AM205 (ddl) | 172027..173031 [+], 1005 | 56416508 | D-alanine--D-alanine ligase | | |
| 3 | AM206 (ftsQ) | 173024..173797 [+], 774 | 56416509 | cell division protein | | |
| 4 | AM207 (ntrX) | 174038..175438 [+], 1401 | 56416510 | nitrogen assimilation regulatory protein | | |
| 5 | AM210 (orfX) | 175825..176214 [+], 390 | 56416511 | hypothetical protein | VirB2 | |
| 6 | AM215 (gor) | 177599..179011 [-], 1413 | 56416512 | glutathione reductase | | |
| |
| Region 4: 259077..259331 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM299 | 254937..255290 [-], 354 | 56416573 | hypothetical protein | | |
| 2 | AM301 (rpsD) | 255537..256073 [+], 537 | 56416574 | 30S ribosomal protein S4 | | |
| 3 | AM302 | 256490..257638 [-], 1149 | 56416575 | hypothetical protein | | |
| 4 | AM303 | 257698..258471 [-], 774 | 56416576 | hypothetical protein | | |
| 5 | AM305 (pgpA) | 258486..259010 [-], 525 | 56416577 | phosphatidylglycerophosphatase A | | |
| 6 | AM306 | 259077..259331 [-], 255 | 56416578 | hypothetical protein | VirB7 | |
| 7 | AM308 (tig) | 259718..261118 [+], 1401 | 56416579 | trigger factor | | |
| 8 | AM309 (clpP) | 261123..261770 [+], 648 | 56416580 | ATP-dependent Clp protease proteolytic subunit | | |
| 9 | AM310 (clpX) | 261780..262997 [+], 1218 | 56416581 | ATP-dependent protease ATP-binding subunit ClpX | | |
| |
| Region 5: 659399..663547 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM715 | 658530..658817 [-], 288 | 56416858 | hypothetical protein | | |
| 2 | AM717 (orfX) | 659399..659770 [-], 372 | 56416859 | hypothetical protein | VirB2 | |
| 3 | AM721 (orfY) | 662843..663076 [+], 234 | 56416860 | hypothetical protein | VirB2 | |
| 4 | AM723 (orfX) | 663149..663547 [+], 399 | 56416861 | hypothetical protein | VirB2 | |
| 5 | AM730 (typA) | 664842..666701 [-], 1860 | 56416862 | GTP-binding protein | | |
| |
| Region 6: 734837..75243 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM804 (ispH) | 730871..731899 [+], 1029 | 56416914 | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | | |
| 2 | AM805 (dut) | 732101..732544 [+], 444 | 56416915 | deoxyuridine 5'triphosphate nucleotidohydrolase | | |
| 3 | AM807 (murF) | 732591..733982 [-], 1392 | 56416916 | UDP-N-acetylmuramyl-tripeptide synthetase | | |
| 4 | AM809 | 734205..734822 [+], 618 | 56416917 | hypothetical protein | | |
| 5 | AM810 | 734837..739900 [-], 5064 | 56416918 | hypothetical protein | VirB6 | |
| 6 | AM811 | 739863..743978 [-], 4116 | 56416919 | hypothetical protein | VirB6 | |
| 7 | AM812 | 743990..747049 [-], 3060 | 56416920 | hypothetical protein | VirB6 | |
| 8 | AM813 (virB6) | 747102..749738 [-], 2637 | 56416921 | VirB6 protein | VirB6 | |
| 9 | AM814 (virB4) | 749731..752139 [-], 2409 | 56416922 | type IV secretion system ATPase VirB4 | VirB4 | |
| 10 | AM815 (virB3) | 752143..752439 [-], 297 | 56416923 | type IV secretion system protein VirB3 | VirB3 | |
| 11 | AM816 (sodB) | 752533..753204 [-], 672 | 56416924 | Fe superoxide dismutase | | |
| 12 | AM818 (pstA) | 753368..754753 [+], 1386 | 56416925 | phosphate ABC transporter | | |
| 13 | AM820 (lipA) | 754976..755917 [-], 942 | 56416927 | lipoyl synthase | | |
| 14 | AM821 | 756603..758225 [-], 1623 | 56416928 | hypothetical protein | | |
| 15 | AM823 | 758458..762411 [-], 3954 | 56416929 | hypothetical protein | | |
| 16 | AM824 (pycA) | 762503..764491 [-], 1989 | 56416930 | propionyl-CoA carboxylase subunit alpha | | |
| 17 | AM825 (sfhB) | 764861..765844 [+], 984 | 56416931 | sfhb-like protein | | |
| 18 | AM826 (rpmB) | 765847..766251 [-], 405 | 56416932 | 50S ribosomal protein L28 | | |
| 19 | AM827 (priA) | 766755..768743 [-], 1989 | 56416933 | primosomal protein N' | | |
| 20 | AM829 | 768961..769884 [-], 924 | 56416934 | hypothetical protein | | |
| 21 | AM830 | 770010..770813 [-], 804 | 56416935 | hypothetical protein | | |
| 22 | AM832 | 770910..772622 [-], 1713 | 56416936 | hypothetical protein | | |
| 23 | AM833 | 772749..773276 [-], 528 | 56416937 | hypothetical protein | | |
| 24 | AM836 | 773664..774524 [-], 861 | 56416938 | hypothetical protein | | |
| 25 | AM837 (era) | 774500..775393 [-], 894 | 56416939 | GTP-binding protein Era | | |
| 26 | AM839 (ffh) | 775488..776843 [-], 1356 | 56416940 | signal recognition particle protein | | |
| 27 | AM840 (nuoL3) | 776836..778386 [-], 1551 | 56416941 | monovalent cation/H+ antiporter subunit D | | |
| 28 | AM841 | 778461..778937 [-], 477 | 56416942 | hypothetical protein | | |
| 29 | AM842 (dnaK) | 779164..781101 [-], 1938 | 56416943 | molecular chaperone DnaK | | |
| 30 | AM844 (rne) | 781301..783160 [-], 1860 | 56416944 | ribonuclease E | | |
| 31 | AM846 | 783255..783581 [-], 327 | 56416945 | monovalent cation/H+ antiporter subunit C | | |
| 32 | AM847 | 783668..784144 [-], 477 | 56416946 | monovalent cation/H+ antiporter subunit B | | |
| 33 | AM848 | 784101..784670 [-], 570 | 56416947 | monovalent cation/H+ antiporter subunit B | | |
| 34 | AM849 | 784671..784970 [-], 300 | 56416948 | monovalent cation/H+ antiporter subunit G | | |
| 35 | AM851 | 784977..785231 [-], 255 | 56416949 | monovalent cation/H+ antiporter subunit F | | |
| 36 | AM852 (tktA) | 785249..787234 [-], 1986 | 56416950 | transketolase | | |
| 37 | AM853 | 787250..788854 [-], 1605 | 56416951 | hypothetical protein | | |
| 38 | AM854 | 789000..789710 [+], 711 | 56416952 | hypothetical protein | | |
| 39 | AM856 (purA) | 790004..791341 [+], 1338 | 56416953 | adenylosuccinate synthetase | | |
| 40 | AM858 | 791345..792331 [-], 987 | 56416954 | hypothetical protein | | |
| 41 | AM859 | 792369..792863 [-], 495 | 56416955 | hypothetical protein | | |
| 42 | AM860 (nrdA) | 792908..794728 [-], 1821 | 56416956 | ribonucleotide-diphosphate reductase subunit alpha | | |
| 43 | AM861 (ispA) | 795296..796171 [+], 876 | 56416957 | geranyltranstransferase (farnesyl-diphosphate synthase) | | |
| 44 | AM862 | 796220..797116 [+], 897 | 56416958 | hypothetical protein | | |
| 45 | AM863 (thiO) | 797146..798135 [-], 990 | 56416959 | thiamine biosynthesis oxidoreductase | | |
| 46 | AM865 (dnaB) | 798415..799890 [-], 1476 | 56416960 | replicative DNA helicase | | |
| 47 | AM867 | 799944..800618 [-], 675 | 56416961 | hypothetical protein | | |
| 48 | AM868 (ssnA) | 800782..801246 [+], 465 | 56416962 | cytosine deaminase | | |
| 49 | AM869 (fabH) | 801378..802346 [-], 969 | 56416963 | 3-oxoacyl-ACP synthase | | |
| 50 | AM870 (plsX) | 802346..803389 [-], 1044 | 56416964 | glycerol-3-phosphate acyltransferase PlsX | | |
| 51 | AM870.5 (rpmF) | 803389..803565 [-], 177 | 56416965 | 50S ribosomal protein L32 | | |
| 52 | AM871 (tgt) | 803779..805041 [+], 1263 | 56416966 | queuine tRNA-ribosyltransferase | | |
| 53 | AM872 (pstB) | 805237..806061 [-], 825 | 56416967 | phosphate ABC transporter ATP-binding protein | | |
| 54 | AM873 (dapB) | 806001..806843 [-], 843 | 56416968 | dihydrodipicolinate reductase | | |
| 55 | AM875 | 806956..807441 [-], 486 | 56416969 | hypothetical protein | | |
| 56 | AM876 (ubiA) | 807454..808353 [-], 900 | 56416970 | 4-hydroxybenzoate polyprenyltransferase | | |
| 57 | AM878 (aaaP) | 808427..809686 [-], 1260 | 56416971 | appendage-associated protein | | |
| 58 | AM879 | 809841..811124 [-], 1284 | 56416972 | hypothetical protein | | |
| 59 | AM880 | 811267..812109 [-], 843 | 56416973 | hypothetical protein | | |
| 60 | AM880.5 (rpmH) | 812267..812401 [+], 135 | 56416974 | 50S ribosomal protein L34 | | |
| 61 | AM881 | 812407..812766 [+], 360 | 56416975 | hypothetical protein | | |
| 62 | AM882 | 812779..814098 [+], 1320 | 56416976 | hypothetical protein | | |
| 63 | AM884 | 814157..816403 [-], 2247 | 56416977 | hypothetical protein | | |
| 64 | AM885 (pheT) | 816685..819048 [-], 2364 | 56416978 | phenylalanyl-tRNA synthetase subunit beta | | |
| 65 | AM886 (rplQ) | 819052..819456 [-], 405 | 56416979 | 50S ribosomal protein L17 | | |
| 66 | AM887 (rpoA) | 819497..820597 [-], 1101 | 56416980 | DNA-directed RNA polymerase subunit alpha | | |
| 67 | AM888 (rpsK) | 820618..820992 [-], 375 | 161544985 | 30S ribosomal protein S11 | | |
| 68 | AM890 (rpsM) | 821002..821373 [-], 372 | 56416982 | 30S ribosomal protein S13 | | |
| 69 | AM891 (adk) | 821437..822087 [-], 651 | 56416983 | adenylate kinase | | |
| 70 | AM892 (secY) | 822069..823370 [-], 1302 | 56416984 | preprotein translocase subunit SecY | | |
| 71 | AM893 (rplO) | 823394..823834 [-], 441 | 56416985 | 50S ribosomal protein L15 | | |
| 72 | AM894 (rpsE) | 823848..824372 [-], 525 | 56416986 | 30S ribosomal protein S5 | | |
| 73 | AM895 (rplR) | 824383..824760 [-], 378 | 56416987 | 50S ribosomal protein L18 | | |
| 74 | AM896 (rplF) | 824762..825307 [-], 546 | 56416988 | 50S ribosomal protein L6 | | |
| 75 | AM897 (rpsH) | 825319..825717 [-], 399 | 56416989 | 30S ribosomal protein S8 | | |
| 76 | AM898 (rpsN) | 825735..825983 [-], 249 | 56416990 | small ribosomal protein S14 | | |
| 77 | AM899 (rplE) | 826052..826591 [-], 540 | 56416991 | 50S ribosomal protein L5 | | |
| 78 | AM900 (rplX) | 826592..826909 [-], 318 | 56416992 | 50S ribosomal protein L24 | | |
| 79 | AM901 (rplN) | 826906..827265 [-], 360 | 56416993 | 50S ribosomal protein L14 | | |
| 80 | AM903 (rpsQ) | 827278..827511 [-], 234 | 56416994 | 30S ribosomal protein S17 | | |
| 81 | AM904 (rpmC) | 827501..827707 [-], 207 | 56416995 | 50S ribosomal protein L29 | | |
| 82 | AM905 (rplP) | 827715..828131 [-], 417 | 161544984 | 50S ribosomal protein L16 | | |
| 83 | AM906 (rpsC) | 828140..828775 [-], 636 | 56416997 | 30S ribosomal protein S3 | | |
| 84 | AM907 (rplV) | 828779..829117 [-], 339 | 56416998 | 50S ribosomal protein L22 | | |
| 85 | AM908 (rpsS) | 829121..829402 [-], 282 | 56416999 | 30S ribosomal protein S19 | | |
| 86 | AM909 (rplB) | 829408..830241 [-], 834 | 56417000 | 50S ribosomal protein L2 | | |
| 87 | AM910 (rplW) | 830238..830546 [-], 309 | 56417001 | 50S ribosomal protein L23 | | |
| 88 | AM911 (rplD) | 830521..831147 [-], 627 | 56417002 | 50S ribosomal protein L4 | | |
| 89 | AM912 (rplC) | 831165..831863 [-], 699 | 56417003 | 50S ribosomal protein L3 | | |
| 90 | AM913 (rpsJ) | 831865..832182 [-], 318 | 56417004 | 30S ribosomal protein S10 | | |
| 91 | AM914 (tuf) | 832189..833370 [-], 1182 | 56417005 | elongation factor Tu | | |
| 92 | AM915 | 833746..834531 [+], 786 | 56417006 | hypothetical protein | | |
| 93 | AM916 (cmk) | 834518..835195 [+], 678 | 56417007 | cytidylate kinase | | |
| 94 | AM917 (rpsA) | 835260..836939 [+], 1680 | 56417008 | 30S ribosomal protein S1 | | |
| 95 | AM918 (sppA) | 836943..837824 [+], 882 | 56417009 | protease IV | | |
| 96 | AM919 | 838073..838570 [+], 498 | 56417010 | hypothetical protein | | |
| 97 | AM920 | 838571..839008 [+], 438 | 56417011 | hypothetical protein | | |
| 98 | AM921 | 839324..840277 [+], 954 | 56417012 | hypothetical protein | | |
| 99 | AM922 (hemH) | 840581..841633 [+], 1053 | 56417013 | ferrochelatase | | |
| 100 | AM923 | 841636..842700 [-], 1065 | 56417014 | hypothetical protein | | |
| 101 | AM924 | 842956..843282 [-], 327 | 56417015 | hypothetical protein | | |
| 102 | AM925 (hflK) | 843819..844742 [-], 924 | 56417016 | HFLK protein | | |
| 103 | AM926 | 844805..845653 [-], 849 | 56417017 | hypothetical protein | | |
| 104 | AM927 | 845751..848354 [+], 2604 | 56417018 | hypothetical protein | | |
| 105 | AM928 (qor) | 848679..849662 [-], 984 | 56417019 | quinone oxidoreductase | | |
| 106 | AM929 | 849768..852170 [-], 2403 | 56417020 | hypothetical protein | | |
| 107 | AM931 (folP) | 852504..853310 [+], 807 | 56417021 | dihydropteroate synthase | | |
| 108 | AM933 (carB) | 853769..857077 [+], 3309 | 56417022 | carbamoyl phosphate synthase large subunit | | |
| 109 | AM936 | 858293..858685 [-], 393 | 56417023 | hypothetical protein | | |
| 110 | AM937 (fumC) | 858850..860238 [-], 1389 | 56417024 | fumarate hydratase | | |
| 111 | AM938 | 860408..861058 [+], 651 | 56417025 | hypothetical protein | | |
| 112 | AM939 (pyrC) | 861059..862423 [+], 1365 | 56417026 | dihydroorotase | | |
| 113 | AM940 (glmM) | 862557..863915 [-], 1359 | 56417027 | phosphoglucosamine mutase | | |
| 114 | AM941 (lipB) | 863952..864596 [-], 645 | 56417028 | lipoyltransferase | | |
| 115 | AM942 (radC) | 864687..865379 [-], 693 | 56417029 | DNA repair protein | | |
| 116 | AM943 (groES) | 865600..865884 [+], 285 | 161544983 | co-chaperonin GroES | | |
| 117 | AM944 (groEL) | 865932..867581 [+], 1650 | 56417031 | chaperonin GroEL | | |
| 118 | AM947 | 868196..868927 [-], 732 | 56417032 | hypothetical protein | | |
| 119 | AM949 (clpB) | 869149..871767 [+], 2619 | 56417033 | ATP-dependent Clp protease ATP-binding subunit | | |
| 120 | AM950 | 871879..873540 [-], 1662 | 56417034 | hypothetical protein | | |
| 121 | AM952 | 873771..874763 [-], 993 | 56417035 | hypothetical protein | | |
| 122 | AM953 | 874868..875608 [-], 741 | 56417036 | hypothetical protein | | |
| 123 | AM955 | 875813..876211 [+], 399 | 56417037 | hypothetical protein | | |
| 124 | AM956 (pepA) | 876183..877787 [+], 1605 | 56417038 | leucyl aminopeptidase | | |
| 125 | AM957 | 877803..878447 [+], 645 | 56417039 | hypothetical protein | | |
| 126 | AM959 | 878594..880069 [-], 1476 | 56417040 | hypothetical protein | | |
| 127 | AM960 (purQ) | 880211..880993 [-], 783 | 56417041 | phosphoribosylformylglycinamidine synthase | | |
| 128 | AM961 (gidA) | 881074..882987 [+], 1914 | 56417042 | tRNA uridine 5-carboxymethylaminomethyl modification protein GidA | | |
| 129 | AM964 (glpX) | 883300..884265 [-], 966 | 56417043 | fructose 1,6-bisphosphatase II | | |
| 130 | AM965 | 884352..885053 [+], 702 | 56417044 | hypothetical protein | | |
| 131 | AM967 | 885213..885668 [+], 456 | 56417045 | hypothetical protein | | |
| 132 | AM968 | 885769..886053 [+], 285 | 56417046 | hypothetical protein | | |
| 133 | AM969 (bioB) | 886040..887014 [+], 975 | 56417047 | biotin synthase | | |
| 134 | AM973 (purL) | 887742..890792 [+], 3051 | 56417048 | phosphoribosylformylglycinamidine synthase | | |
| 135 | AM974 | 890823..891029 [+], 207 | 56417049 | hypothetical protein | | |
| 136 | AM975 | 891166..891870 [+], 705 | 56417050 | hypothetical protein | | |
| 137 | AM976 | 891926..892141 [-], 216 | 56417051 | hypothetical protein | | |
| 138 | AM977 | 892150..892698 [-], 549 | 56417052 | hypothetical protein | | |
| 139 | AM979 | 892797..894077 [-], 1281 | 56417053 | hypothetical protein | | |
| 140 | AM980 | 894252..894620 [+], 369 | 56417054 | hypothetical protein | | |
| 141 | AM981 | 894865..895755 [+], 891 | 56417055 | hypothetical protein | | |
| 142 | AM982 | 895993..896646 [-], 654 | 56417056 | hypothetical protein | | |
| 143 | AM983 (purM) | 896760..897815 [+], 1056 | 56417057 | phosphoribosylaminoimidazole synthetase | | |
| 144 | AM984 (xerC) | 897807..898757 [-], 951 | 56417058 | integrase/recombinase ripx | | |
| 145 | AM985 (gpsA) | 898926..900095 [+], 1170 | 56417059 | gylcerol-3-phosphate dehydrogenase | | |
| 146 | AM987 (omp15) | 900092..900547 [+], 456 | 56417060 | outer membrane protein 15 | | |
| 147 | AM989 (orfX) | 900783..901157 [-], 375 | 56417061 | hypothetical protein | VirB2 | |
| 148 | AM990 (orfY) | 901157..901468 [-], 312 | 56417062 | hypothetical protein | VirB2 | |
| 149 | AM992 (rho) | 901852..903234 [-], 1383 | 56417063 | transcription termination factor Rho | | |
| 150 | AM997 | 903504..903842 [-], 339 | 56417064 | hypothetical protein | | |
| 151 | AM998 (hslV) | 904024..904593 [+], 570 | 56417065 | ATP-dependent protease peptidase subunit | | |
| 152 | AM999 (hslU) | 904590..906038 [+], 1449 | 56417066 | ATP-dependent protease ATP-binding subunit HslU | | |
| 153 | AM1000 (ubiE) | 906042..906785 [+], 744 | 56417067 | ubiquinone/menaquinone biosynthesis methyltransferase | | |
| 154 | AM1001 (metS) | 906786..908153 [-], 1368 | 56417068 | methionyl-tRNA synthetase | | |
| 155 | AM1005 (lysC) | 908599..909837 [+], 1239 | 56417069 | aspartate kinase | | |
| 156 | AM1006 | 909871..910086 [-], 216 | 56417070 | hypothetical protein | | |
| 157 | AM1007 (coxB) | 910094..910924 [+], 831 | 56417071 | cytochrome C oxidase subunit II | | |
| 158 | AM1008 (coxA) | 911021..912586 [+], 1566 | 56417072 | cytochrome C oxidase subunit I | | |
| 159 | AM1009 (ctaB) | 912596..913501 [+], 906 | 56417073 | protoheme IX farnesyltransferase | | |
| 160 | AM1010 | 913554..914480 [-], 927 | 56417074 | hypothetical protein | | |
| 161 | AM1011 (purD) | 914624..915886 [+], 1263 | 56417075 | phosphoribosylamine--glycine ligase | | |
| 162 | AM1012 (yajC) | 915917..916423 [+], 507 | 56417076 | preprotein translocase subunit | | |
| 163 | AM1013 (glmS) | 916492..918318 [+], 1827 | 56417077 | glucosamine--fructose-6-phosphate aminotransferase | | |
| 164 | AM1014 (dnaN) | 918685..919824 [-], 1140 | 56417078 | DNA polymerase III subunit beta | | |
| 165 | AM1015 (rnd) | 920045..920659 [-], 615 | 56417079 | ribonuclease D | | |
| 166 | AM1016 (czcR) | 920724..921596 [-], 873 | 56417080 | transcriptional activator protein | | |
| 167 | AM1018 | 921897..922544 [-], 648 | 56417081 | hypothetical protein | | |
| 168 | AM1020 (ppa) | 923388..923915 [-], 528 | 56417082 | inorganic phosphatase protein | | |
| 169 | AM1021 | 924979..925203 [-], 225 | 56417083 | hypothetical protein | | |
| 170 | AM1022 (rpsI) | 925139..925600 [-], 462 | 56417084 | 30S ribosomal protein S9 | | |
| 171 | AM1023 (rplM) | 925572..926084 [-], 513 | 56417085 | 50S ribosomal protein L13 | | |
| 172 | AM1024 (tolC) | 926120..927517 [+], 1398 | 56417086 | outer membrane protein | | |
| 173 | AM1025 | 927536..928129 [+], 594 | 56417087 | hypothetical protein | | |
| 174 | AM1027 | 928257..929288 [-], 1032 | 56417088 | hypothetical protein | | |
| 175 | AM1028 (prsA) | 929392..930171 [+], 780 | 56417089 | phosphoribosylpyrophosphate synthetase | | |
| 176 | AM1030 | 930364..930684 [+], 321 | 56417090 | hypothetical protein | | |
| 177 | AM1032 (acnA) | 930720..933356 [-], 2637 | 161544982 | aconitate hydratase | | |
| 178 | AM1035 (apaG) | 933744..934196 [-], 453 | 56417092 | ApaG protein | | |
| 179 | AM1036 (purK) | 934272..935342 [+], 1071 | 56417093 | phosphoribosylaminoimidazole carboxylase | | |
| 180 | AM1037 | 935472..937625 [-], 2154 | 56417094 | hypothetical protein | | |
| 181 | AM1041 | 938242..939717 [+], 1476 | 56417095 | hypothetical protein | | |
| 182 | AM1046 | 940029..941474 [+], 1446 | 56417096 | hypothetical protein | | |
| 183 | AM1048 | 941505..945434 [-], 3930 | 56417097 | hypothetical protein | | |
| 184 | AM1050 | 945638..949768 [-], 4131 | 56417098 | hypothetical protein | | |
| 185 | AM1051 | 950058..955655 [-], 5598 | 56417099 | hypothetical protein | | |
| 186 | AM1053 (virB4) | 956209..958638 [-], 2430 | 56417100 | VirB4 protein | VirB4 | |
| 187 | AM1054 | 958771..959145 [-], 375 | 56417101 | hypothetical protein | VirB2 | |
| 188 | AM1055 | 959418..959795 [-], 378 | 56417102 | hypothetical protein | VirB2 | |
| 189 | AM1056 | 960582..961172 [-], 591 | 56417103 | hypothetical protein | VirB2 | |
| 190 | AM1057 | 961182..961730 [-], 549 | 56417104 | hypothetical protein | | |
| 191 | AM1058 | 961959..962324 [+], 366 | 56417105 | hypothetical protein | | |
| 192 | AM1059 (orfY) | 962297..962566 [+], 270 | 56417106 | hypothetical protein | VirB2 | |
| 193 | AM1061 (orfX) | 962633..963028 [+], 396 | 56417107 | hypothetical protein | VirB2 | |
| 194 | AM1063 (msp3) | 963365..966196 [+], 2832 | 56417108 | major surface protein 3 | | |
| 195 | AM1064 (hflK) | 966443..967555 [+], 1113 | 56417109 | hflK protein | | |
| 196 | AM1065 (hflC) | 967557..968429 [+], 873 | 56417110 | hflC protein | | |
| 197 | AM1066 | 968432..969895 [+], 1464 | 56417111 | hypothetical protein | | |
| 198 | AM1067 | 969888..970502 [+], 615 | 56417112 | hypothetical protein | | |
| 199 | AM1069 (rnc) | 970489..971187 [+], 699 | 56417113 | ribonuclease III | | |
| 200 | AM1070 (cox11) | 971218..971841 [+], 624 | 56417114 | cytochrome C oxidase assembly protein | | |
| 201 | AM1073 | 971851..972633 [-], 783 | 56417115 | hypothetical protein | | |
| 202 | AM1076 (ana29) | 972972..973766 [-], 795 | 56417116 | hypothetical protein | | |
| 203 | AM1077 | 973899..974120 [+], 222 | 56417117 | hypothetical protein | | |
| 204 | AM1078 | 974177..975052 [-], 876 | 56417118 | hypothetical protein | | |
| 205 | AM1079 | 975249..976583 [-], 1335 | 56417119 | hypothetical protein | | |
| 206 | AM1080 | 976596..978017 [-], 1422 | 56417120 | hypothetical protein | | |
| 207 | AM1081 (lspA) | 977980..978492 [-], 513 | 56417121 | lipoprotein signal peptidase | | |
| 208 | AM1082 (ribF) | 978571..979482 [-], 912 | 56417122 | riboflavin kinase / FAD synthetase protein | | |
| 209 | AM1083 (grxC1) | 979550..979792 [+], 243 | 56417123 | glutaredoxin | | |
| 210 | AM1084 (hemK) | 979944..980801 [-], 858 | 56417124 | protein-(glutamine-N5) methyltransferase | | |
| 211 | AM1086 (map) | 980982..981779 [-], 798 | 56417125 | methionine aminopeptidase | | |
| 212 | AM1087 (sucB) | 981783..983096 [-], 1314 | 56417126 | dihydrolipoamide acetyltransferase component | | |
| 213 | AM1088 | 983127..983516 [-], 390 | 56417127 | hypothetical protein | | |
| 214 | AM1089 (panC) | 983746..984645 [-], 900 | 56417128 | pantoate--beta-alanine ligase | | |
| 215 | AM1091 | 984734..985876 [+], 1143 | 56417129 | hypothetical protein | | |
| 216 | AM1092 (dsbD) | 985863..987092 [-], 1230 | 56417130 | cytochrome C biogenesis protein | | |
| 217 | AM1093 | 987136..987468 [-], 333 | 56417131 | hypothetical protein | | |
| 218 | AM1094 | 987506..988609 [+], 1104 | 56417132 | hypothetical protein | | |
| 219 | AM1096 (oma87) | 988615..990954 [+], 2340 | 56417133 | outer membrane protein | | |
| 220 | AM1097 | 990941..991501 [+], 561 | 56417134 | hypothetical protein | | |
| 221 | AM1098 (fabZ) | 991507..991950 [+], 444 | 56417135 | (3R)-hydroxymyristoyl-ACP dehydratase | | |
| 222 | AM1099 (purH) | 991965..993506 [+], 1542 | 56417136 | phosphoribosylaminoimidazolecarboxamide formyltransferase (AICAR transformylase) | | |
| 223 | AM1100 (pgsA) | 993515..994057 [-], 543 | 56417137 | CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase | | |
| 224 | AM1103 | 994766..996070 [+], 1305 | 56417138 | hypothetical protein | | |
| 225 | AM1104 (slt) | 996098..998122 [+], 2025 | 56417139 | soluble lytic murein transglycosylase precursor | | |
| 226 | AM1105 (surf1) | 998129..998815 [-], 687 | 56417140 | surfeit locus protein 1 | | |
| 227 | AM1108 | 999373..1001166 [+], 1794 | 56417141 | hypothetical protein | | |
| 228 | AM1109 | 1001259..1002611 [-], 1353 | 56417142 | hypothetical protein | | |
| 229 | AM1110 (atpB) | 1002680..1003483 [+], 804 | 56417143 | F0F1 ATP synthase subunit A | | |
| 230 | AM1111 | 1003567..1004193 [-], 627 | 56417144 | hypothetical protein | | |
| 231 | AM1112 (atpE) | 1004848..1005072 [+], 225 | 161544981 | F0F1 ATP synthase subunit C | | |
| 232 | AM1113 | 1005079..1005579 [+], 501 | 56417146 | hypothetical protein | | |
| 233 | AM1114 | 1005576..1006073 [+], 498 | 56417147 | hypothetical protein | | |
| 234 | AM1115 (ftsA) | 1006198..1007472 [-], 1275 | 56417148 | cell division protein FtsA | | |
| 235 | AM1116 (trkH) | 1007504..1009075 [-], 1572 | 56417149 | potassium uptake protein | | |
| 236 | AM1117 (mesJ) | 1009146..1010651 [+], 1506 | 56417150 | cell cycle protein | | |
| 237 | AM1119 (ftsH) | 1010664..1012496 [+], 1833 | 56417151 | cell division protein | | |
| 238 | AM1120 (secA) | 1012689..1015307 [+], 2619 | 161544980 | preprotein translocase subunit SecA | | |
| 239 | AM1121 | 1015158..1015364 [-], 207 | 56417153 | hypothetical protein | | |
| 240 | AM1122 | 1015383..1016132 [-], 750 | 56417154 | hypothetical protein | | |
| 241 | AM1123 (czcD) | 1016136..1017089 [-], 954 | 56417155 | cation efflux system protein | | |
| 242 | AM1125 (ccmB) | 1017256..1017918 [-], 663 | 56417156 | heme exporter protein B | | |
| 243 | AM1126 (dksA) | 1018019..1018432 [+], 414 | 56417157 | dnaK suppressor protein | | |
| 244 | AM1128 (dtbS) | 1018760..1019443 [-], 684 | 56417158 | dethiobiotin synthase | | |
| 245 | AM1129 (ftsY) | 1019519..1020436 [+], 918 | 56417159 | cell division protein | | |
| 246 | AM1131 (icd) | 1020711..1022177 [+], 1467 | 56417160 | isocitrate dehydrogenase | | |
| 247 | AM1133 (recJ) | 1022302..1024062 [+], 1761 | 56417161 | single-stranded-DNA-specific exonuclease | | |
| 248 | AM1135 | 1024037..1024336 [-], 300 | 56417162 | hypothetical protein | | |
| 249 | AM1136 (pcnB) | 1024527..1025786 [+], 1260 | 56417163 | poly (A) polymerase | | |
| 250 | AM1137 (ndk) | 1025923..1026363 [-], 441 | 56417164 | nucleoside diphosphate kinase | | |
| 251 | AM1138 | 1026592..1027155 [+], 564 | 56417165 | hypothetical protein | | |
| 252 | AM1139 (omp1) | 1027570..1028562 [+], 993 | 56417166 | outer membrane protein 1 | | |
| 253 | AM1140 (opag3) | 1028727..1029665 [+], 939 | 56417167 | msp2 operon associated gene 3 | | |
| 254 | AM1141 | 1028745..1029848 [-], 1104 | 56417168 | hypothetical protein | | |
| 255 | AM1142 (opag2) | 1029680..1030591 [+], 912 | 56417169 | msp2 operon associated gene 2 | | |
| 256 | AM1143 (opag1) | 1030615..1031007 [+], 393 | 56417170 | msp2 operon associated gene 1 | | |
| 257 | AM1144 (msp2) | 1031020..1032243 [+], 1224 | 56417171 | major surface protein 2 | | |
| 258 | AM1145 | 1032455..1032949 [-], 495 | 56417172 | hypothetical protein | | |
| 259 | AM1146 | 1032906..1033490 [+], 585 | 56417173 | hypothetical protein | | |
| 260 | AM1149 (orfX) | 1034507..1034896 [-], 390 | 56417174 | hypothetical protein | VirB2 | |
| 261 | AM1156 (omp2) | 1038392..1039126 [-], 735 | 56417175 | outer membrane protein 2 | | |
| 262 | AM1159 (omp3) | 1039207..1039857 [-], 651 | 56417176 | outer membrane protein 3 | | |
| 263 | AM1161 | 1039827..1040150 [+], 324 | 56417177 | hypothetical protein | | |
| 264 | AM1164 (omp4) | 1040420..1041637 [+], 1218 | 56417178 | outer membrane protein 4 | | |
| 265 | AM1165 | 1041615..1041863 [+], 249 | 56417179 | hypothetical protein | | |
| 266 | AM1166 (omp5) | 1041982..1043100 [+], 1119 | 56417180 | outer membrane protein 5 | | |
| 267 | AM1168 | 1043468..1044283 [-], 816 | 56417181 | hypothetical protein | | |
| 268 | AM1169 (ccmF) | 1044334..1046256 [-], 1923 | 56417182 | cytochrome C-type biogenesis protein | | |
| 269 | AM1170 (valS) | 1046615..1049086 [+], 2472 | 56417183 | valyl-tRNA synthetase | | |
| 270 | AM1171 (smpB) | 1049982..1050431 [-], 450 | 56417184 | SsrA-binding protein | | |
| 271 | AM1172 (ribA) | 1050434..1051066 [-], 633 | 56417185 | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | |
| 272 | AM1173 (greA) | 1051085..1051609 [-], 525 | 56417186 | transcription elongation factor greA | | |
| 273 | AM1174 (atpA) | 1051606..1053129 [-], 1524 | 56417187 | F0F1 ATP synthase subunit alpha | | |
| 274 | AM1175 (atpH) | 1053149..1053721 [-], 573 | 56417188 | ATP synthase subunit delta | | |
| 275 | AM1179 | 1054297..1054548 [-], 252 | 56417189 | hypothetical protein | | |
| 276 | AM1180 | 1054552..1055790 [-], 1239 | 56417190 | hypothetical protein | | |
| 277 | AM1181 | 1055787..1056539 [-], 753 | 56417191 | hypothetical protein | | |
| 278 | AM1183 | 1056544..1057770 [-], 1227 | 56417192 | hypothetical protein | | |
| 279 | AM1185 | 1058051..1059100 [+], 1050 | 56417193 | hypothetical protein | | |
| 280 | AM1186 | 1059095..1060324 [-], 1230 | 56417194 | hypothetical protein | | |
| 281 | AM1187 (gltA) | 1060680..1061933 [-], 1254 | 56417195 | citrate synthase | | |
| 282 | AM1188 (guaA) | 1061970..1063559 [-], 1590 | 56417196 | GMP synthase | | |
| 283 | AM1189 | 1063676..1063906 [-], 231 | 56417197 | hypothetical protein | | |
| 284 | AM1191 | 1064038..1064433 [+], 396 | 56417198 | hypothetical protein | | |
| 285 | AM1192 (pdhC) | 1065049..1066350 [+], 1302 | 56417199 | branched-chain alpha-keto acid dehydrogenase E2 subunit | | |
| 286 | AM1193 | 1066507..1066761 [-], 255 | 56417200 | hypothetical protein | | |
| 287 | AM1194 (fbaB) | 1066669..1067568 [+], 900 | 161544979 | fructose-bisphosphate aldolase | | |
| 288 | AM1196 (pbpA1) | 1067822..1069627 [-], 1806 | 56417202 | penicillin-binding protein | | |
| 289 | AM1197 (secF) | 1069608..1070543 [-], 936 | 56417203 | preprotein translocase subunit SecF | | |
| 290 | AM1200 | 1070689..1071069 [+], 381 | 56417204 | hypothetical protein | | |
| 291 | AM1202 (hemA) | 1071072..1072304 [-], 1233 | 56417205 | 5-aminolevulinate synthase | | |
| 292 | AM1205 (tyrS) | 1072474..1073718 [-], 1245 | 56417206 | tyrosyl-tRNA synthetase | | |
| 293 | AM1206 (glnA) | 1074028..1075434 [+], 1407 | 56417207 | glutamine synthetase | | |
| 294 | AM1207 (mraY) | 1075445..1076473 [+], 1029 | 56417208 | phospho-N-acetylmuramoyl-pentapeptide-transferase | | |
| 295 | AM1208 (ispB) | 1076483..1077466 [+], 984 | 56417209 | octaprenyl-diphosphate synthase | | |
| 296 | AM1209 | 1077541..1079622 [-], 2082 | 56417210 | hypothetical protein | | |
| 297 | AM1210 (mkl) | 1079749..1080468 [-], 720 | 56417211 | ribonucleotide transport ATP-binding protein | | |
| 298 | AM1212 | 1080473..1081252 [-], 780 | 56417212 | hypothetical protein | | |
| 299 | AM1213 (rpe) | 1081271..1082053 [-], 783 | 56417213 | ribulose-phosphate 3-epimerase | | |
| 300 | AM1214 (polA) | 1082130..1084724 [-], 2595 | 56417214 | DNA polymerase I | | |
| 301 | AM1216 | 1085541..1085927 [+], 387 | 56417215 | hypothetical protein | | |
| 302 | AM1217 (argF) | 1085950..1086858 [+], 909 | 56417216 | ornithine carbamoyltransferase | | |
| 303 | AM1218 (recF) | 1086868..1087983 [+], 1116 | 56417217 | recombination protein F | | |
| 304 | AM1219 (omp6) | 1087980..1088441 [-], 462 | 56417218 | outer membrane protein 6 | | |
| 305 | AM1220 (omp7) | 1088481..1089671 [-], 1191 | 56417219 | outer membrane protein 7 | | |
| 306 | AM1221 (omp8) | 1089736..1090935 [-], 1200 | 56417220 | outer membrane protein 8 | | |
| 307 | AM1222 (omp9) | 1090999..1092210 [-], 1212 | 56417221 | outer membrane protein 9 | | |
| 308 | AM1223 (omp10) | 1092273..1093535 [-], 1263 | 56417222 | outer membrane protein 10 | | |
| 309 | AM1225 | 1093976..1094518 [-], 543 | 56417223 | hypothetical protein | | |
| 310 | AM1226 | 1094876..1095514 [+], 639 | 56417224 | hypothetical protein | | |
| 311 | AM1228 | 1095979..1096902 [-], 924 | 56417225 | hypothetical protein | | |
| 312 | AM1229 (mviN) | 1097053..1098417 [-], 1365 | 56417226 | virulence factor MVIN | | |
| 313 | AM1231 (lgt) | 1098643..1099500 [+], 858 | 56417227 | prolipoprotein diacylglyceryl transferase | | |
| 314 | AM1232 | 1099740..1099955 [+], 216 | 56417228 | hypothetical protein | | |
| 315 | AM1233 (rpsT) | 1100204..1100494 [-], 291 | 161544978 | 30S ribosomal protein S20 | | |
| 316 | AM1235 (plsC) | 1100613..1101338 [-], 726 | 56417230 | 1-acyl-sn-glycerol-3-phosphate acyltransferase | | |
| 317 | AM1236 (def) | 1101370..1101927 [+], 558 | 56417231 | N-formylmethionylaminoacyl-tRNA deformylase | | |
| 318 | AM1237 | 1102174..1103046 [+], 873 | 56417232 | hypothetical protein | | |
| 319 | AM1238 | 1103188..1103916 [+], 729 | 56417233 | hypothetical protein | | |
| 320 | AM1239 | 1104007..1104234 [-], 228 | 56417234 | hypothetical protein | | |
| 321 | AM1240 | 1104293..1104499 [-], 207 | 56417235 | hypothetical protein | | |
| 322 | AM1241 | 1104735..1105244 [-], 510 | 56417236 | hypothetical protein | | |
| 323 | AM1242 | 1105289..1106398 [-], 1110 | 56417237 | hypothetical protein | | |
| 324 | AM1243 (uvrB) | 1106811..1108766 [-], 1956 | 56417238 | excinuclease ABC subunit B | | |
| 325 | AM1245 (secD) | 1108831..1110336 [+], 1506 | 56417239 | preprotein translocase subunit SecD | | |
| 326 | AM1248 | 1110826..1112649 [+], 1824 | 56417240 | hypothetical protein | | |
| 327 | AM1251 (orfY) | 1113315..1113548 [+], 234 | 56417241 | hypothetical protein | VirB2 | |
| 328 | AM1253 (orfX) | 1113621..1114010 [+], 390 | 56417242 | hypothetical protein | VirB2 | |
| 329 | AM1254 | 1114075..1115559 [-], 1485 | 56417243 | hypothetical protein | | |
| 330 | AM1255 (omp11) | 1115669..1116658 [-], 990 | 56417244 | outer membrane protein 11 | | |
| 331 | AM1257 (omp12) | 1116852..1117709 [-], 858 | 56417245 | outer membrane protein 12 | | |
| 332 | AM1258 (omp13) | 1117917..1118975 [-], 1059 | 56417246 | outer membrane protein 13 | | |
| 333 | AM1261 (ftsZ) | 1119261..1120514 [+], 1254 | 56417247 | cell division protein FtsZ | | |
| 334 | AM1264 | 1120891..1121355 [+], 465 | 56417248 | hypothetical protein | | |
| 335 | AM1266 (parA) | 1121429..1122244 [+], 816 | 56417249 | chromosome partitioning protein | | |
| 336 | AM1267 (parB) | 1122241..1123068 [+], 828 | 56417250 | chromosome partitioning protein | | |
| 337 | AM1269 (rimM) | 1123080..1123652 [+], 573 | 56417251 | 16S rRNA-processing protein RimM | | |
| 338 | AM1270 (trmD) | 1123649..1124356 [+], 708 | 56417252 | tRNA (guanine-N(1)-)-methyltransferase | | |
| 339 | AM1271 (rplS) | 1124349..1124753 [+], 405 | 56417253 | 50S ribosomal protein L19 | | |
| 340 | AM1272 (cinA) | 1124760..1125281 [-], 522 | 56417254 | competence / damage-inducible protein | | |
| 341 | AM1273 (thrS) | 1125347..1127257 [+], 1911 | 56417255 | threonyl-tRNA synthetase | | |
| 342 | AM1274 (infC) | 1127287..1127820 [+], 534 | 56417256 | translation initiation factor IF-3 | | |
| 343 | AM1275 | 1128227..1128682 [-], 456 | 56417257 | hypothetical protein | | |
| 344 | AM1276 | 1128686..1129387 [-], 702 | 56417258 | hypothetical protein | | |
| 345 | AM1277 (asd) | 1129505..1130515 [-], 1011 | 56417259 | aspartate-semialdehyde dehydrogenase | | |
| 346 | AM1278 | 1130517..1130825 [-], 309 | 56417260 | hypothetical protein | | |
| 347 | AM1280 (dnaZ) | 1130831..1132255 [-], 1425 | 56417261 | DNA polymerase III subunit gamma | | |
| 348 | AM1283 (gapB) | 1132441..1133445 [-], 1005 | 56417262 | NAD(P)-dependent glyceraldehyde 3-phosphate dehydrogenase | | |
| 349 | AM1285 | 1133834..1134778 [+], 945 | 56417263 | hypothetical protein | | |
| 350 | AM1286 | 1134794..1135450 [+], 657 | 56417264 | isoprenoid biosynthesis protein | | |
| 351 | AM1287 (ftsW) | 1135475..1136632 [-], 1158 | 56417265 | cell division protein | | |
| 352 | AM1288 (metK) | 1136614..1137816 [-], 1203 | 56417266 | S-adenosylmethionine synthetase | | |
| 353 | AM1289 (ubiH) | 1138038..1139204 [+], 1167 | 56417267 | 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase | | |
| 354 | AM1292 | 1139216..1139824 [-], 609 | 56417268 | hypothetical protein | | |
| 355 | AM1294 (glyQ) | 1140100..1140951 [+], 852 | 56417269 | glycyl-tRNA synthetase subunit alpha | | |
| 356 | AM1295 (glyS) | 1140953..1143037 [+], 2085 | 56417270 | glycyl-tRNA synthetase subunit beta | | |
| 357 | AM1296 (dnaJ) | 1143119..1144258 [+], 1140 | 56417271 | chaperone protein DnaJ | | |
| 358 | AM1297 (nadC) | 1144277..1145095 [-], 819 | 56417272 | nicotinate-nucleotide pyrophosphorylase | | |
| 359 | AM1298 | 1145154..1145750 [+], 597 | 56417273 | membrane protein | | |
| 360 | AM1299 (ruvC) | 1145742..1146239 [-], 498 | 56417274 | holliday (crossover) junction endodeoxyribonuclease | | |
| 361 | AM1300 (coxC) | 1146258..1147076 [-], 819 | 56417275 | cytochrome C oxidase subunit III | | |
| 362 | AM1301 (hemE) | 1147146..1147943 [-], 798 | 56417276 | uroporphyrinogen decarboxylase | | |
| 363 | AM1303 | 1148564..1149445 [+], 882 | 56417277 | hypothetical protein | | |
| 364 | AM1304 | 1149432..1149950 [-], 519 | 56417278 | hypothetical protein | | |
| 365 | AM1305 | 1149934..1151130 [-], 1197 | 161544977 | hypothetical protein | | |
| 366 | AM1306 | 1151233..1152033 [+], 801 | 56417280 | hypothetical protein | | |
| 367 | AM1307 (nadA) | 1152053..1153009 [+], 957 | 56417281 | quinolinate synthetase | | |
| 368 | AM1309 (fdxA) | 1153031..1153402 [-], 372 | 56417282 | ferredoxin II | | |
| 369 | AM1310 (ilvC) | 1153528..1154499 [+], 972 | 56417283 | ketol-acid reductoisomerase | | |
| 370 | AM1311 (infC) | 1154526..1154783 [-], 258 | 56417284 | translation initiation factor IF-3 | | |
| 371 | AM1312 (virD4) | 1154957..1157377 [-], 2421 | 56417285 | type IV secretion system protein VirB8 | VirD4 | |
| 372 | AM1313 (virB11) | 1157425..1158438 [-], 1014 | 56417286 | type IV secretion system ATPase VirB11 | VirB11 | |
| 373 | AM1314 (virB10) | 1158480..1159817 [-], 1338 | 56417287 | VirB10 protein | VirB10 | |
| 374 | AM1315 (virB9) | 1159825..1160664 [-], 840 | 56417288 | VirB9 protein | VirB9 | |
| 375 | AM1316 (virB8) | 1160651..1161340 [-], 690 | 56417289 | VirB8 protein | VirB8 | |
| 376 | AM1317 (ribA) | 1161459..1162592 [-], 1134 | 56417290 | GTP cyclohydrolase II | | |
| 377 | AM1319 | 1162709..1163950 [-], 1242 | 56417291 | hypothetical protein | | |
| 378 | AM1320 | 1163892..1164419 [-], 528 | 56417292 | hypothetical protein | | |
| 379 | AM1323 (dapF) | 1164480..1165313 [-], 834 | 56417293 | diaminopimelate epimerase | | |
| 380 | AM1324 | 1165325..1166041 [-], 717 | 56417294 | hypothetical protein | | |
| 381 | AM1325 | 1166183..1166611 [-], 429 | 56417295 | hypothetical protein | | |
| 382 | AM1326 (pgk) | 1167047..1168204 [+], 1158 | 56417296 | phosphoglycerate kinase | | |
| 383 | AM1327 (xseA) | 1168396..1169589 [-], 1194 | 56417297 | exodeoxyribonuclease large subunit | | |
| 384 | AM1328 | 1169564..1170232 [-], 669 | 56417298 | hypothetical protein | | |
| 385 | AM1331 (dapD) | 1170411..1171247 [+], 837 | 161544976 | 2,3,4,5-tetrahydropyridine-2,6-carboxylate N-succinyltransferase | | |
| 386 | AM1332 (trmE) | 1171248..1172579 [+], 1332 | 56417300 | tRNA modification GTPase TrmE | | |
| 387 | AM1335 (dfp) | 1172736..1173293 [-], 558 | 56417301 | phosphopantothenoylcysteine decarboxylase | | |
| 388 | AM1336 (recG) | 1173303..1175402 [+], 2100 | 56417302 | ATP-dependent DNA helicase | | |
| 389 | AM1337 | 1175585..1176565 [+], 981 | 56417303 | hypothetical protein | | |
| 390 | AM1338 (purF) | 1176700..1178109 [-], 1410 | 56417304 | amidophosphoribosyltransferase | | |
| 391 | AM1339 (pth) | 1178260..1178820 [-], 561 | 56417305 | peptidyl-tRNA hydrolase | | |
| 392 | AM1340 (rplY) | 1178825..1179481 [-], 657 | 56417306 | 50S ribosomal protein L25/general stress protein Ctc | | |
| 393 | AM1341 | 1179523..1179741 [-], 219 | 56417307 | hypothetical protein | | |
| 394 | AM1346 | 1183255..1184004 [-], 750 | 56417308 | hypothetical protein | | |
| 395 | AM1347 (dapE) | 1183943..1185145 [-], 1203 | 56417309 | succinyl-diaminopimelate desuccinylase | | |
| 396 | AM1348 (kefB) | 1185315..1187006 [-], 1692 | 56417310 | glutathione-regulated potassium-efflux system protein | | |
| 397 | AM1349 | 1187020..1187583 [-], 564 | 56417311 | hypothetical protein | | |
| 398 | AM1350 | 1187811..1188257 [-], 447 | 56417312 | hypothetical protein | | |
| 399 | AM1351 (pdhB) | 1188388..1189413 [-], 1026 | 56417313 | pyruvate dehydrogenase subunit beta | | |
| 400 | AM1352 | 1189478..1192339 [-], 2862 | 56417314 | hypothetical protein | | |
| 401 | AM1353 (tldD) | 1192852..1194273 [+], 1422 | 56417315 | tldD protein | | |
| 402 | AM1354 | 1194357..1194593 [-], 237 | 56417316 | hypothetical protein | | |
| 403 | AM1355 | 1194703..1195797 [-], 1095 | 56417317 | GTP-dependent nucleic acid-binding protein EngD | | |
| 404 | AM1356 (ispF) | 1195978..1196568 [-], 591 | 56417318 | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | |
| 405 | AM1357 (ispD) | 1196596..1197306 [-], 711 | 56417319 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | | |
| 406 | AM1359 | 1..1905 [+], 1905 | 56416371 | hypothetical protein | | |
| 407 | AM001 (purE) | 1862..2362 [-], 501 | 56416372 | phosphoribosylaminoimidazole carboxylase catalytic subunit | | |
| 408 | AM002 | 2355..2957 [-], 603 | 56416373 | hypothetical protein | | |
| 409 | AM005 (rodA) | 3113..4183 [+], 1071 | 56416374 | rod shape determining protein | | |
| 410 | AM006 | 4258..4554 [+], 297 | 56416375 | hypothetical protein | | |
| 411 | AM007 | 4551..4925 [+], 375 | 56416376 | hypothetical protein | | |
| 412 | AM008 | 5187..6119 [-], 933 | 56416377 | hypothetical protein | | |
| 413 | AM009 | 6209..6619 [-], 411 | 56416378 | hypothetical protein | | |
| 414 | AM010 (murC) | 6921..7544 [+], 624 | 56416379 | UDP-N-acetylmuramate-alanine ligase | | |
| 415 | AM011 (murC) | 7532..8368 [+], 837 | 56416380 | UDP-N-acetylmuramate-alanine ligase | | |
| 416 | AM012 (trpS) | 8395..9384 [-], 990 | 56416381 | tryptophanyl-tRNA synthetase | | |
| 417 | AM013 (grpE) | 9381..9875 [-], 495 | 56416382 | GRPE protein | | |
| 418 | AM015 (ribD) | 10097..11212 [+], 1116 | 56416383 | riboflavin biosynthesis protein | | |
| 419 | AM018 (pyrG) | 11486..13168 [-], 1683 | 56416384 | CTP synthetase | | |
| 420 | AM019 (secG) | 13181..13510 [-], 330 | 56416385 | preprotein translocase subunit SecG | | |
| 421 | AM020 | 13686..13946 [+], 261 | 56416386 | hypothetical protein | | |
| 422 | AM021 (xerD) | 13939..14877 [+], 939 | 56416387 | integrase/recombinase | | |
| 423 | AM023 (phnL) | 15075..15749 [-], 675 | 56416388 | ATP transporter ATP-binding protein | | |
| 424 | AM024 (maeB) | 16020..18272 [+], 2253 | 56416389 | malic enzyme | | |
| 425 | AM025 | 18446..21166 [+], 2721 | 56416390 | hypothetical protein | | |
| 426 | AM026 | 21386..22900 [+], 1515 | 56416391 | monovalent cation/H+ antiporter subunit D | | |
| 427 | AM027 | 22915..23301 [-], 387 | 56416392 | hypothetical protein | | |
| 428 | AM030 (orfX) | 23496..23882 [-], 387 | 56416393 | hypothetical protein | VirB2 | |
| 429 | AM032 (orfY) | 23955..24188 [-], 234 | 56416394 | hypothetical protein | VirB2 | |
| 430 | AM034 | 25004..27445 [-], 2442 | 56416395 | hypothetical protein | | |
| 431 | AM035 | 27643..27852 [+], 210 | 56416396 | hypothetical protein | | |
| 432 | AM037 (thiG) | 27915..28709 [+], 795 | 56416397 | thiazole synthase | | |
| 433 | AM038 | 28807..29247 [-], 441 | 56416398 | hypothetical protein | | |
| 434 | AM039 (gltX) | 29429..30754 [+], 1326 | 56416399 | glutamyl-tRNA synthetase | | |
| 435 | AM041 | 31186..32391 [+], 1206 | 56416400 | hypothetical protein | | |
| 436 | AM042 | 32410..32967 [+], 558 | 56416401 | hypothetical protein | | |
| 437 | AM044 (orfX) | 33143..33541 [-], 399 | 56416402 | hypothetical protein | VirB2 | |
| 438 | AM046 (orfY) | 33614..33847 [-], 234 | 56416403 | hypothetical protein | VirB2 | |
| 439 | AM051 (rho) | 36761..38125 [-], 1365 | 56416404 | transcription termination factor Rho | | |
| 440 | AM052 | 38234..38851 [-], 618 | 56416405 | hypothetical protein | | |
| 441 | AM053 (rpmI) | 39269..39469 [+], 201 | 56416406 | 50S ribosomal protein L35 | | |
| 442 | AM054 (rplT) | 39496..39882 [+], 387 | 56416407 | 50S ribosomal protein L20 | | |
| 443 | AM055 (pheS) | 39845..40900 [+], 1056 | 56416408 | phenylalanyl-tRNA synthetase subunit alpha | | |
| 444 | AM056 | 40991..41749 [+], 759 | 56416409 | hypothetical protein | | |
| 445 | AM058 (glmU) | 41930..43216 [+], 1287 | 56416410 | UDP-N-acetylglucosamine pyrophosphorylase | | |
| 446 | AM059 (rpsP) | 43369..43638 [-], 270 | 56416411 | 30S ribosomal protein S16 | | |
| 447 | AM060 (proP1) | 43929..45215 [+], 1287 | 56416412 | proline/betaine transport protein | | |
| 448 | AM063 (orfY) | 47834..48067 [+], 234 | 56416413 | hypothetical protein | VirB2 | |
| 449 | AM065 (orfX) | 48140..48535 [+], 396 | 56416414 | hypothetical protein | VirB2 | |
| 450 | AM069 (fpbA) | 49429..50469 [+], 1041 | 56416415 | iron binding protein FbpA precursor | | |
| 451 | AM070 | 50470..50649 [+], 180 | 56416416 | hypothetical protein | | |
| 452 | AM071 | 50899..54888 [-], 3990 | 56416417 | hypothetical protein | | |
| 453 | AM072 | 55153..61272 [+], 6120 | 56416418 | hypothetical protein | | |
| 454 | AM073 (nrdR) | 61443..61898 [-], 456 | 56416419 | NrdR family transcriptional regulator | | |
| 455 | AM074 | 62431..62886 [+], 456 | 56416420 | hypothetical protein | | |
| 456 | AM075 (omp14) | 63055..64269 [+], 1215 | 56416421 | outer membrane protein 14 | | |
| 457 | AM077 (orfX) | 64841..65242 [-], 402 | 56416422 | hypothetical protein | VirB2 | |
| 458 | AM082 (orfX) | 67571..67978 [+], 408 | 56416423 | hypothetical protein | VirB2 | |
| 459 | AM085 (recA) | 68810..69889 [+], 1080 | 56416424 | recombinase A | | |
| 460 | AM087 (pyrE) | 69984..70586 [+], 603 | 56416425 | orotate phosphoribosyltransferase | | |
| 461 | AM088 (thiF) | 70620..71426 [+], 807 | 56416426 | thiamine biosynthesis protein | | |
| 462 | AM090 (msp4) | 71782..72630 [-], 849 | 56416427 | major surface protein 4 | | |
| 463 | AM092 | 75388..76326 [-], 939 | 56416428 | hypothetical protein | | |
| 464 | AM093 | 76411..76731 [-], 321 | 56416429 | hypothetical protein | | |
| 465 | AM094 | 76781..76984 [+], 204 | 56416430 | hypothetical protein | | |
| 466 | AM096 (trxA) | 77419..77766 [-], 348 | 56416431 | thioredoxin | | |
| 467 | AM097 (trbG) | 77775..78587 [-], 813 | 56416432 | conjugal transfer protein trbG precursor | VirB9 | |
| 468 | AM101 (pdhA) | 78838..79956 [-], 1119 | 56416433 | pyruvate dehydrogenase E1 component, subunit alpha precursor | | |
| |
| Region 7: 900783..901468 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM982 | 895993..896646 [-], 654 | 56417056 | hypothetical protein | | |
| 2 | AM983 (purM) | 896760..897815 [+], 1056 | 56417057 | phosphoribosylaminoimidazole synthetase | | |
| 3 | AM984 (xerC) | 897807..898757 [-], 951 | 56417058 | integrase/recombinase ripx | | |
| 4 | AM985 (gpsA) | 898926..900095 [+], 1170 | 56417059 | gylcerol-3-phosphate dehydrogenase | | |
| 5 | AM987 (omp15) | 900092..900547 [+], 456 | 56417060 | outer membrane protein 15 | | |
| 6 | AM989 (orfX) | 900783..901157 [-], 375 | 56417061 | hypothetical protein | VirB2 | |
| 7 | AM990 (orfY) | 901157..901468 [-], 312 | 56417062 | hypothetical protein | VirB2 | |
| 8 | AM992 (rho) | 901852..903234 [-], 1383 | 56417063 | transcription termination factor Rho | | |
| 9 | AM997 | 903504..903842 [-], 339 | 56417064 | hypothetical protein | | |
| 10 | AM998 (hslV) | 904024..904593 [+], 570 | 56417065 | ATP-dependent protease peptidase subunit | | |
| 11 | AM999 (hslU) | 904590..906038 [+], 1449 | 56417066 | ATP-dependent protease ATP-binding subunit HslU | | |
| |
| Region 8: 956209..963028 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM1053 (virB4) | 956209..958638 [-], 2430 | 56417100 | VirB4 protein | VirB4 | |
| 2 | AM1054 | 958771..959145 [-], 375 | 56417101 | hypothetical protein | VirB2 | |
| 3 | AM1055 | 959418..959795 [-], 378 | 56417102 | hypothetical protein | VirB2 | |
| 4 | AM1056 | 960582..961172 [-], 591 | 56417103 | hypothetical protein | VirB2 | |
| 5 | AM1057 | 961182..961730 [-], 549 | 56417104 | hypothetical protein | | |
| 6 | AM1058 | 961959..962324 [+], 366 | 56417105 | hypothetical protein | | |
| 7 | AM1059 (orfY) | 962297..962566 [+], 270 | 56417106 | hypothetical protein | VirB2 | |
| 8 | AM1061 (orfX) | 962633..963028 [+], 396 | 56417107 | hypothetical protein | VirB2 | |
| 9 | AM1063 (msp3) | 963365..966196 [+], 2832 | 56417108 | major surface protein 3 | | |
| 10 | AM1064 (hflK) | 966443..967555 [+], 1113 | 56417109 | hflK protein | | |
| |
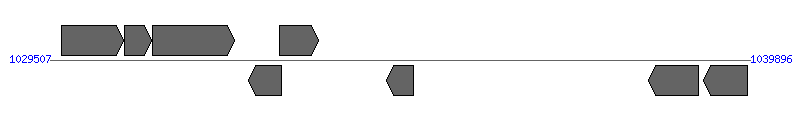
| Region 9: 1034507..1034896 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM1142 (opag2) | 1029680..1030591 [+], 912 | 56417169 | msp2 operon associated gene 2 | | |
| 2 | AM1143 (opag1) | 1030615..1031007 [+], 393 | 56417170 | msp2 operon associated gene 1 | | |
| 3 | AM1144 (msp2) | 1031020..1032243 [+], 1224 | 56417171 | major surface protein 2 | | |
| 4 | AM1145 | 1032455..1032949 [-], 495 | 56417172 | hypothetical protein | | |
| 5 | AM1146 | 1032906..1033490 [+], 585 | 56417173 | hypothetical protein | | |
| 6 | AM1149 (orfX) | 1034507..1034896 [-], 390 | 56417174 | hypothetical protein | VirB2 | |
| 7 | AM1156 (omp2) | 1038392..1039126 [-], 735 | 56417175 | outer membrane protein 2 | | |
| 8 | AM1159 (omp3) | 1039207..1039857 [-], 651 | 56417176 | outer membrane protein 3 | | |
| |
| Region 10: 1113315..1114010 |
 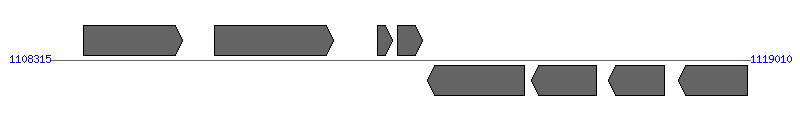 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM1245 (secD) | 1108831..1110336 [+], 1506 | 56417239 | preprotein translocase subunit SecD | | |
| 2 | AM1248 | 1110826..1112649 [+], 1824 | 56417240 | hypothetical protein | | |
| 3 | AM1251 (orfY) | 1113315..1113548 [+], 234 | 56417241 | hypothetical protein | VirB2 | |
| 4 | AM1253 (orfX) | 1113621..1114010 [+], 390 | 56417242 | hypothetical protein | VirB2 | |
| 5 | AM1254 | 1114075..1115559 [-], 1485 | 56417243 | hypothetical protein | | |
| 6 | AM1255 (omp11) | 1115669..1116658 [-], 990 | 56417244 | outer membrane protein 11 | | |
| 7 | AM1257 (omp12) | 1116852..1117709 [-], 858 | 56417245 | outer membrane protein 12 | | |
| 8 | AM1258 (omp13) | 1117917..1118975 [-], 1059 | 56417246 | outer membrane protein 13 | | |
| |
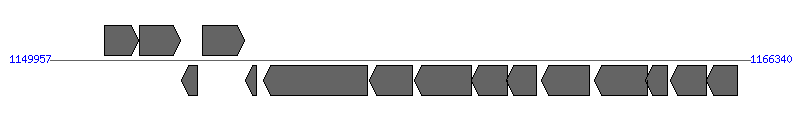
| Region 11: 1154957..1161340 |
  |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | AM1306 | 1151233..1152033 [+], 801 | 56417280 | hypothetical protein | | |
| 2 | AM1307 (nadA) | 1152053..1153009 [+], 957 | 56417281 | quinolinate synthetase | | |
| 3 | AM1309 (fdxA) | 1153031..1153402 [-], 372 | 56417282 | ferredoxin II | | |
| 4 | AM1310 (ilvC) | 1153528..1154499 [+], 972 | 56417283 | ketol-acid reductoisomerase | | |
| 5 | AM1311 (infC) | 1154526..1154783 [-], 258 | 56417284 | translation initiation factor IF-3 | | |
| 6 | AM1312 (virD4) | 1154957..1157377 [-], 2421 | 56417285 | type IV secretion system protein VirB8 | VirD4 | |
| 7 | AM1313 (virB11) | 1157425..1158438 [-], 1014 | 56417286 | type IV secretion system ATPase VirB11 | VirB11 | |
| 8 | AM1314 (virB10) | 1158480..1159817 [-], 1338 | 56417287 | VirB10 protein | VirB10 | |
| 9 | AM1315 (virB9) | 1159825..1160664 [-], 840 | 56417288 | VirB9 protein | VirB9 | |
| 10 | AM1316 (virB8) | 1160651..1161340 [-], 690 | 56417289 | VirB8 protein | VirB8 | |
| 11 | AM1317 (ribA) | 1161459..1162592 [-], 1134 | 56417290 | GTP cyclohydrolase II | | |
| 12 | AM1319 | 1162709..1163950 [-], 1242 | 56417291 | hypothetical protein | | |
| 13 | AM1320 | 1163892..1164419 [-], 528 | 56417292 | hypothetical protein | | |
| 14 | AM1323 (dapF) | 1164480..1165313 [-], 834 | 56417293 | diaminopimelate epimerase | | |
| 15 | AM1324 | 1165325..1166041 [-], 717 | 56417294 | hypothetical protein | | |
| |