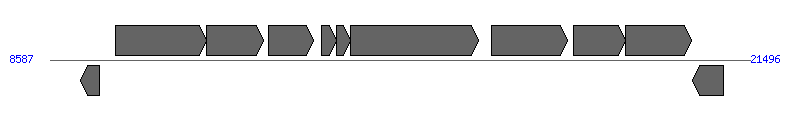The information of T4SS components from NC_011498 |
| Region 1: 13587..16496 |
 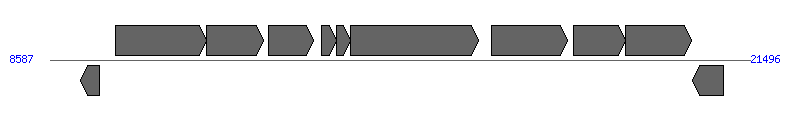 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | HPP12_0009 (groES) | 9151..9507 [-], 357 | 210134210 | co-chaperonin GroES | | |
| 2 | HPP12_0010 (dnaG) | 9795..11474 [+], 1680 | 210134211 | DNA primase | | |
| 3 | HPP12_0011 | 11471..12523 [+], 1053 | 210134212 | argininosuccinate synthase | | |
| 4 | HPP12_0012 | 12615..13442 [+], 828 | 210134213 | hypothetical protein | | |
| 5 | HPP12_0013 (comB2) | 13587..13868 [+], 282 | 210134214 | ComB2 competence protein | ComB2 | |
| 6 | HPP12_0014 (comB3) | 13868..14131 [+], 264 | 210134215 | ComB3 competence protein | ComB3 | |
| 7 | HPP12_0015 (comB4) | 14133..16496 [+], 2364 | 210134216 | ComB4 competence protein | ComB4 | |
| 8 | HPP12_0016 | 16729..18138 [+], 1410 | 210134217 | hypothetical protein | | |
| 9 | HPP12_0017 (cheV-1) | 18238..19203 [+], 966 | 210134218 | chemotaxis signal transduction protein CheV | | |
| 10 | HPP12_0018 (nspC) | 19200..20417 [+], 1218 | 210134219 | carboxynorspermidine decarboxylase | | |
| 11 | HPP12_0019 | 20436..21011 [-], 576 | 210134220 | lipid A 1-phosphatase | | |
| |
| Region 2: 36339..40378 |
 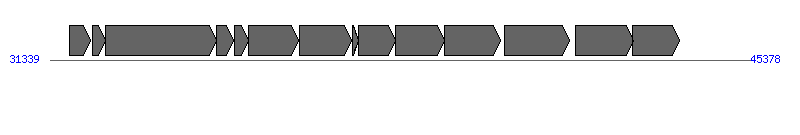 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | HPP12_0027 | 31734..32147 [+], 414 | 210134228 | hypothetical protein | | |
| 2 | HPP12_0028 (clpS) | 32183..32458 [+], 276 | 210134229 | ATP-dependent clp protease adapter protein ClpS | | |
| 3 | HPP12_0029 (clpA) | 32458..34683 [+], 2226 | 210134230 | ATP-dependent clp protease | | |
| 4 | HPP12_0030 (panD) | 34673..35023 [+], 351 | 210134231 | aspartate alpha-decarboxylase | | |
| 5 | HPP12_0031 | 35034..35327 [+], 294 | 210134232 | hypothetical protein | | |
| 6 | HPP12_0032 | 35327..36331 [+], 1005 | 210134233 | hypothetical protein | | |
| 7 | HPP12_0033 (comB6) | 36339..37394 [+], 1056 | 210134234 | ComB6 competence protein | ComB6 | |
| 8 | HPP12_0034 (comb7) | 37410..37523 [+], 114 | 210134235 | Comb7 competence protein | ComB7 | |
| 9 | HPP12_0035 (comB8) | 37520..38263 [+], 744 | 210134236 | ComB8 competence protein | ComB8 | |
| 10 | HPP12_0036 (comB9) | 38263..39249 [+], 987 | 210134237 | ComB9 competence protein | ComB9 | |
| 11 | HPP12_0037 (comB10) | 39242..40378 [+], 1137 | 210134238 | ComB10 competence protein | ComB10 | |
| 12 | HPP12_0038 | 40448..41767 [+], 1320 | 210134239 | bifunctional mannose-6-phosphate isomerase / mannose-1-phosphate guanylyltransferase | | |
| 13 | HPP12_0039 (gmd) | 41888..43033 [+], 1146 | 210134240 | GDP-D-mannose dehydratase | | |
| 14 | HPP12_0040 | 43026..43958 [+], 933 | 210134241 | GDP-fucose synthetase | | |
| |