
| 388 | |
| Xanthomonas campestris pv. campestris str. 8004 | |
| chromosome [Browse all T4SS(s) in this replicon] | |
| NC_007086 | |
| 1956087..1966861 | |
| VirB | |
| effector translocation | |
| Type IVA; Type P | |
T4SS components |
| Component | VirB1 | VirB2 | VirB3 | VirB4 | VirB5 | VirB6 | VirB7 | VirB8 | VirB9 |
| Number | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 |
| Component | VirB10 | VirB11 | VirD4 | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_007086 | ||||||
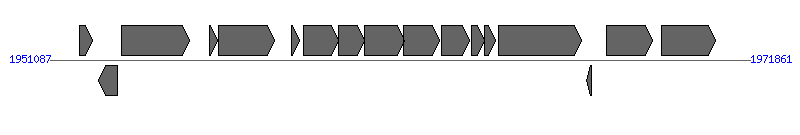 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | XC_1626 | 1951975..1952343 [+], 369 | 66767950 | type IV pilin | ||
| 2 | XC_1627 | 1952524..1953075 [-], 552 | 66767951 | fimbrial biogenesis protein | ||
| 3 | XC_1628 | 1953215..1955236 [+], 2022 | 66767952 | excinuclease ABC subunit B | ||
| 4 | XC_1629 | 1955811..1956053 [+], 243 | 66767953 | hypothetical protein | ||
| 5 | XC_1630 | 1956087..1957760 [+], 1674 | 66767954 | VirD4 protein | VirD4 | |
| 6 | XC_1631 | 1958257..1958499 [+], 243 | 66767955 | hypothetical protein | ||
| 7 | XC_1632 | 1958610..1959653 [+], 1044 | 66767956 | VirB8 protein | VirB8 | |
| 8 | XC_1633 | 1959650..1960417 [+], 768 | 66767957 | VirB9 protein | VirB9 | |
| 9 | XC_1634 | 1960414..1961595 [+], 1182 | 66767958 | VirB10 protein | VirB10 | |
| 10 | XC_1635 | 1961592..1962650 [+], 1059 | 66767959 | VirB11 protein | VirB11 | |
| 11 | XC_1636 | 1962696..1963529 [+], 834 | 66767960 | VirB1 protein | VirB1 | |
| 12 | XC_1637 | 1963592..1963996 [+], 405 | 66767961 | VirB2 protein | VirB2 | |
| 13 | XC_1638 | 1963989..1964300 [+], 312 | 66767962 | VirB3 protein | VirB3 | |
| 14 | XC_1639 | 1964408..1966861 [+], 2454 | 66767963 | VirB4 protein | VirB4 | |
| 15 | XC_1640 | 1967008..1967151 [-], 144 | 66767964 | hypothetical protein | ||
| 16 | XC_1641 | 1967589..1968956 [+], 1368 | 66767965 | IS1478 transposase | ||
| 17 | XC_1642 | 1969222..1970838 [+], 1617 | 66767966 | alpha-glucosidase | ||
Download FASTA format files |
| Proteins Genes |
|
(1) He YQ; Zhang L; Jiang BL; Zhang ZC; Xu RQ; Tang DJ; Qin J; Jiang W; Zhang X; Liao J; Cao JR; Zhang SS; Wei ML; Liang XX; Lu GT; Feng JX; Chen B; Cheng J; Tang JL (2007). Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 8(10):R218. [PudMed:17927820] |
| (2) Qian W; Jia Y; Ren SX; He YQ; Feng JX; Lu LF; Sun Q; Ying G; Tang DJ; Tang H; Wu W; Hao P; Wang L; Jiang BL; Zeng S; Gu WY; Lu G; Rong L; Tian Y; Yao Z; Fu G; Chen B; Fang R; Qiang B; Chen Z; Zhao GP; Tang JL; He C (2005). Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15(6):757-67. [PudMed:15899963] |