Help
Introduction
Natural transformation stands as a fundamental mechanism of horizontal gene transfer (HGT) in bacteria, enables direct uptake and integration of extracellular DNA, driving evolution, adaptation, and antibiotic resistance spread (Johnston, et al. Nat Rev Microbiol, 2014). This multi-stage process (competence regulation, DNA binding, DNA uptake, DNA processing and homologous recombination) is governed by specialized machinery and regulatory genes. Furthermore, several naturally transformable species exhibit DNA uptake specificity, preferentially internalizing fragments containing species-specific motifs (e.g., DUS in Neisseriaceae and USS in Pasteurellaceae) (Frye, et al. PLoS Genet, 2013; Redfield, et al. BMC Evol Biol, 2006).
Mobile genetic elements (MGEs), including insertion sequences (ISs), transposons, genomic islands (GIs), and prophages, exploit transformation for dissemination (e.g., SCCmec acquisition in MRSA; Maree, et al. Nat Commun, 2022). While transformation can purge selfish MGEs (Croucher, et al. PLoS Biol, 2016), MGEs retaliate by suppressing host machinery via extracellular DNA degradation or gene disruption (Dalia, et al. Proc Natl Acad Sci U S A, 2015; Tuffet, et al. Proc Natl Acad Sci USA, 2024), ensuring their persistence.
Key features
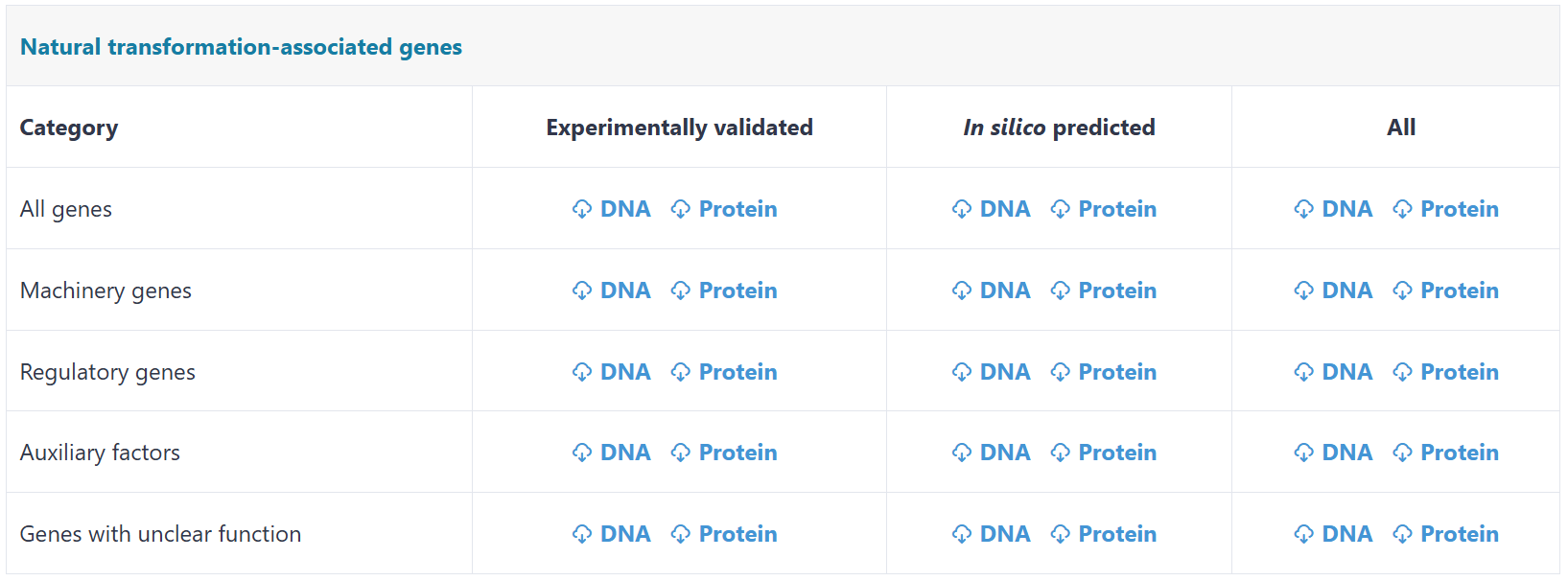
(i) Experimentally validated natural transformation-associated genes. NTDB archives 992 experimentally verified natural transformation-associated genes curated from 782 peer-reviewed publications across 50 prokaryotic species, such as Streptococcus pneumoniae, Vibrio cholerae and Methanococcus maripaludis. Genes were classified into four functional categories based on their roles in natural transformation: (i) Machinery genes (n = 562): the genes directly mediate DNA binding, DNA uptake, DNA processing, or homologous recombination; (ii) Regulatory genes (n = 336): the genes encode regulators operating through transcriptional control, post-translational modification, targeted proteolysis or other regulatory mechanisms to control competence development. Experimentally validated interactions between regulators and their target genes were also archived; (iii) Auxiliary factors (n = 33): comprise restriction-modification (RM) systems or endonucleases modulating transformation efficiency via foreign DNA degradation, and autolysins enabling DNA release through cell lysis; (iv) Genes with unclear function (n = 61): the genes proved to be essential for natural transformation, but the detailed mechanisms remained uncharacterized. Each entry includes annotations such as primary sequence, secondary and tertiary structures, Pfam domains, transmembrane helices, sequence homologs and regulatory network.
(ii) Naturally transformable species without characterized genes. NTDB also catalogs 49 experimentally transformable species whose associated transformation machinery and regulatory genes remain uncharacterized. Natural transformation machinery and regulatory genes in these species were predicted and annotated.
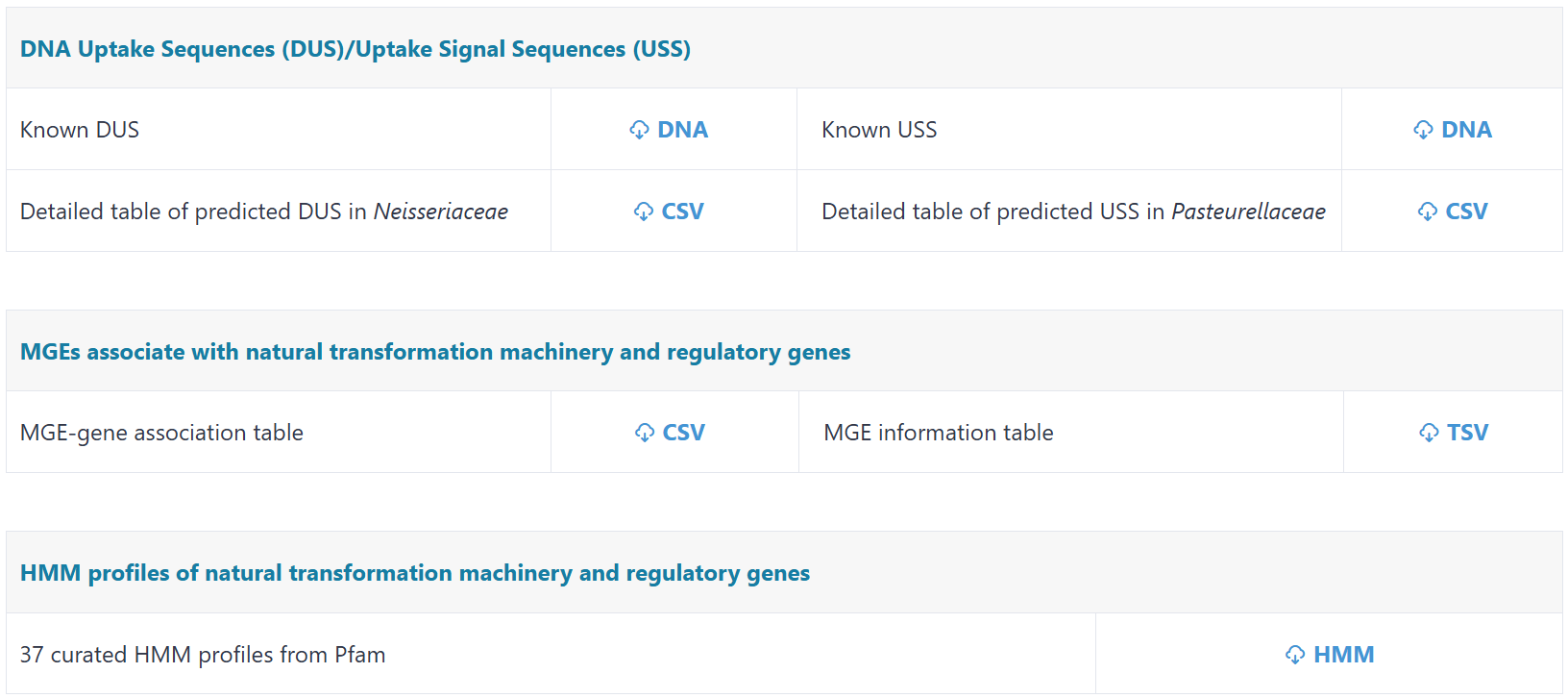
(iii) DNA uptake motifs (DUS/USS). Experimentally validated DNA uptake motifs are cataloged, with computational prediction tools applied across relevant genomes.
(iv) Genome-scale prediction of natural transformation machinery and regulatory genes. A total of 514,235 predicted genes, including 360,601 machinery genes and 153,634 regulatory genes, were identified in the chromosomes of 48,764 prokaryotic complete genomes (48,136 bacterial, 628 archaeal) from the NCBI RefSeq database by BLASTp with Ha-value (identities * coverage) ≥ 0.48.
(i) Identification of MGEs associated with natural transformation machinery and regulatory genes. By utilizing VRprofile2 (Wang, et al. Nucleic Acids Research, 2022), MGEs were identified in 48,764 prokaryotic chromosomes from RefSeq and 115 chromosomes harboring the experimentally validated natural transformation machinery and regulatory genes. By screening both experimentally verified and predicted natural transformation machinery and regulatory genes located completely within or in the 1 kb flanking regions of MGEs, we identified 42,850 associations connecting 41,319 genes (29,610 machinery genes; 11,709 regulatory genes) to 31,941 distinct MGEs.
(ii) Interactive exploration on the website. For each gene involved in natural transformation, the related MGE are listed in Browse tables and displayed in Detailed Information web pages by gene structure plots.
(i) Genome prediction. Upload prokaryotic genomes to predict natural transformation machinery and regulatory genes using NTDB's reference database (BLASTp-based).
(ii) BLAST search. Identify machinery and regulatory genes in query sequences via BLASTp (protein queries), BLASTx (nucleotide-translated queries) or BLASTn (nucleotide queries).
(iii) HMMER search. Screen query protein sequences against 37 curated Pfam HMM profiles related to natural transformation machinery genes for functional domain detection.
(iv) DNA uptake motif search. Predict DNA uptake motifs (DUS/USS) in user-submitted DNA sequences using experimentally validated DNA uptake motifs as references.
Key functions

Usage
How to use NTDB?

The web-based NTDB database contains several major parts: Home, Browse, Statistics, Tools, Download, References, Help, and a search field. Users can browse and download resources according to their needs on the corresponding pages.

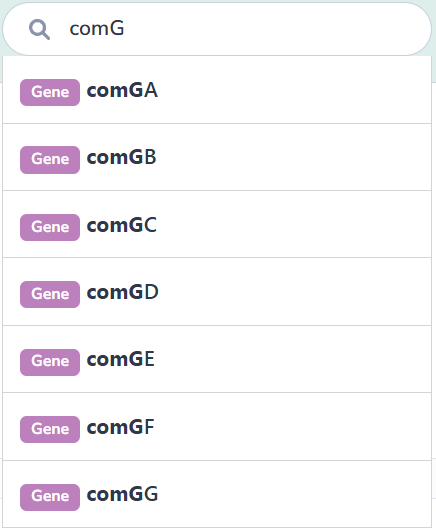
On the "Browse" page
NTDB provides two complementary browsing modalities:
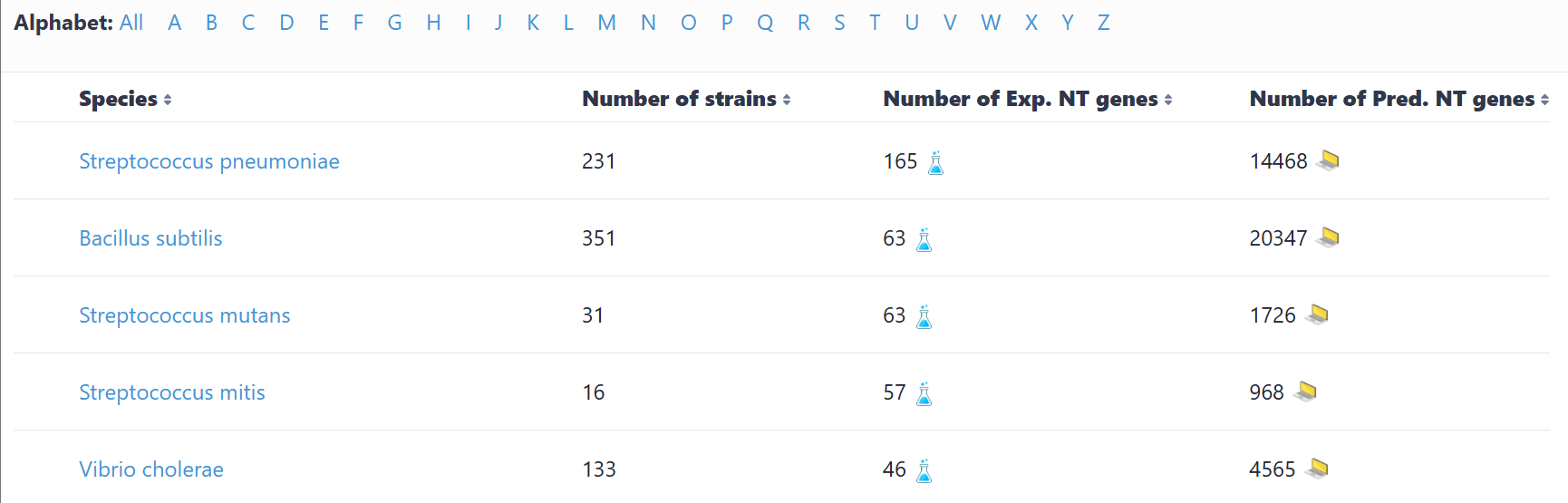
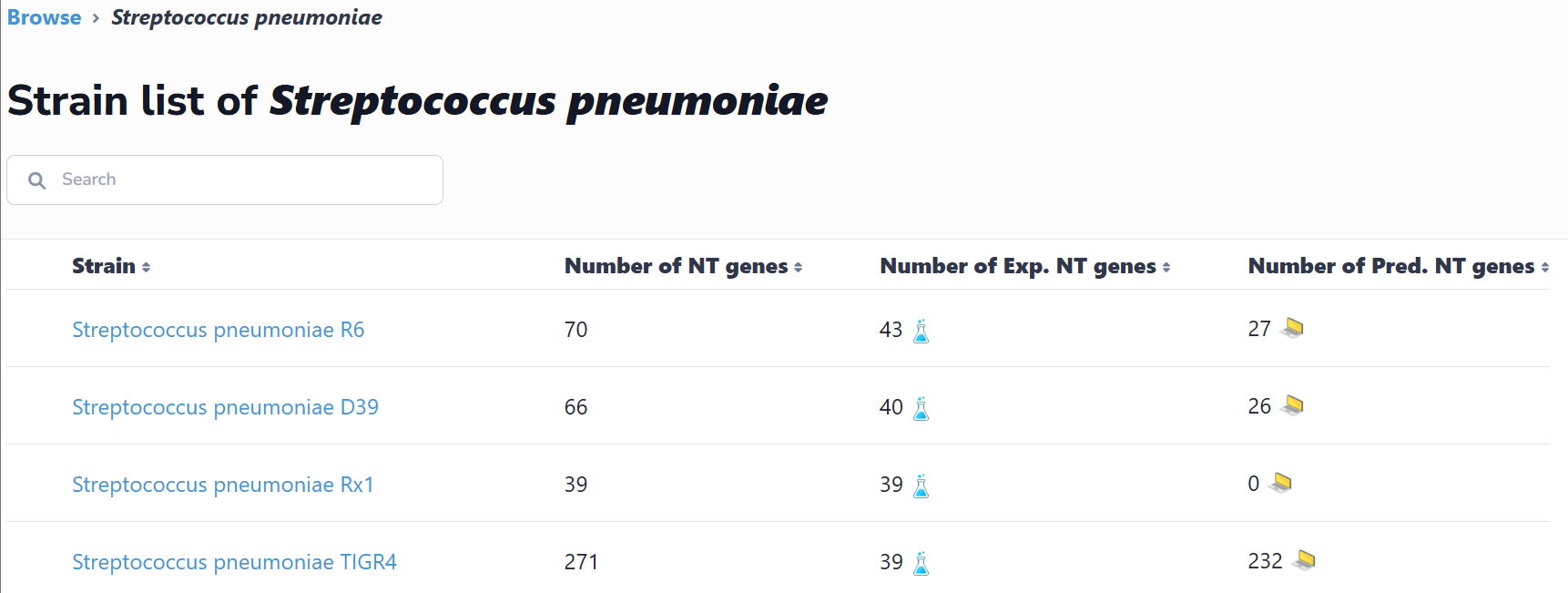
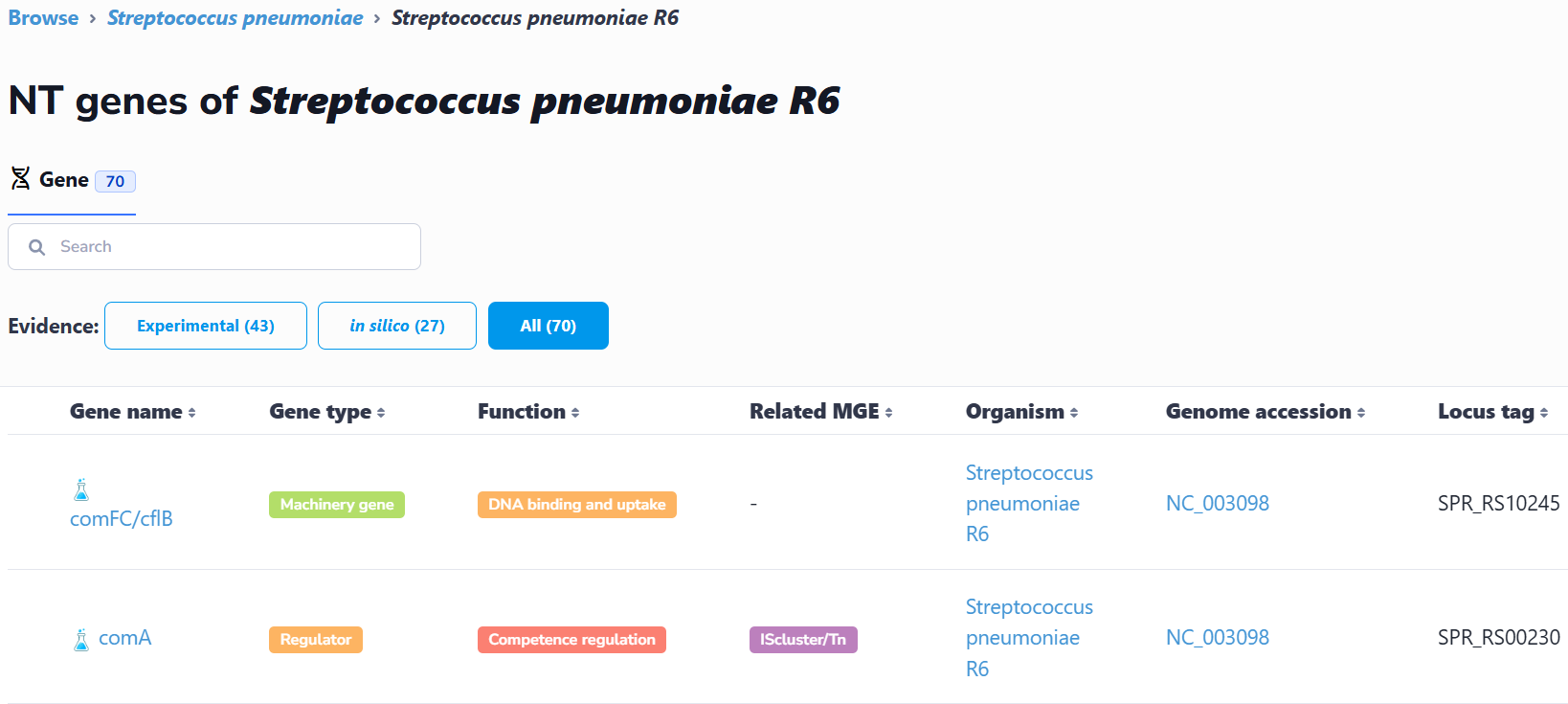
(1) Browse by species
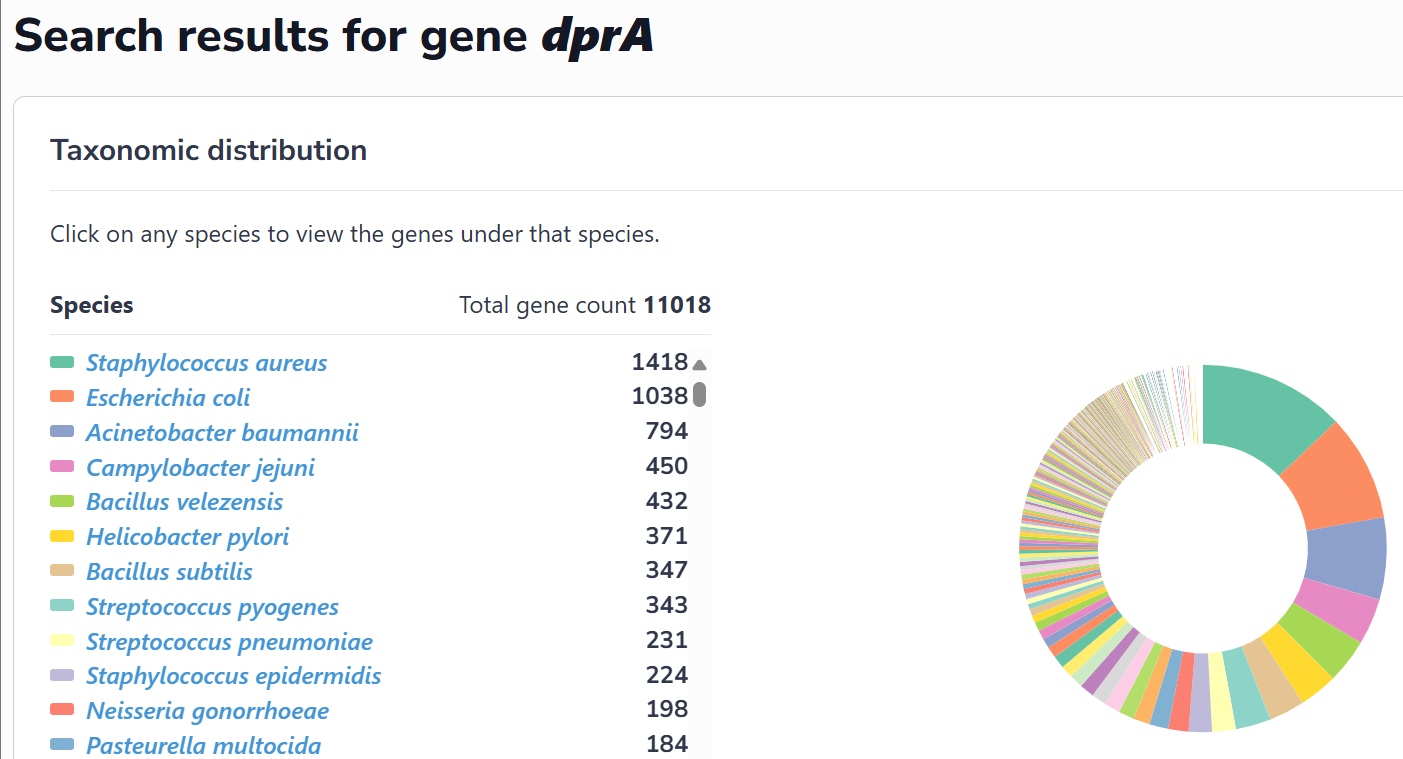
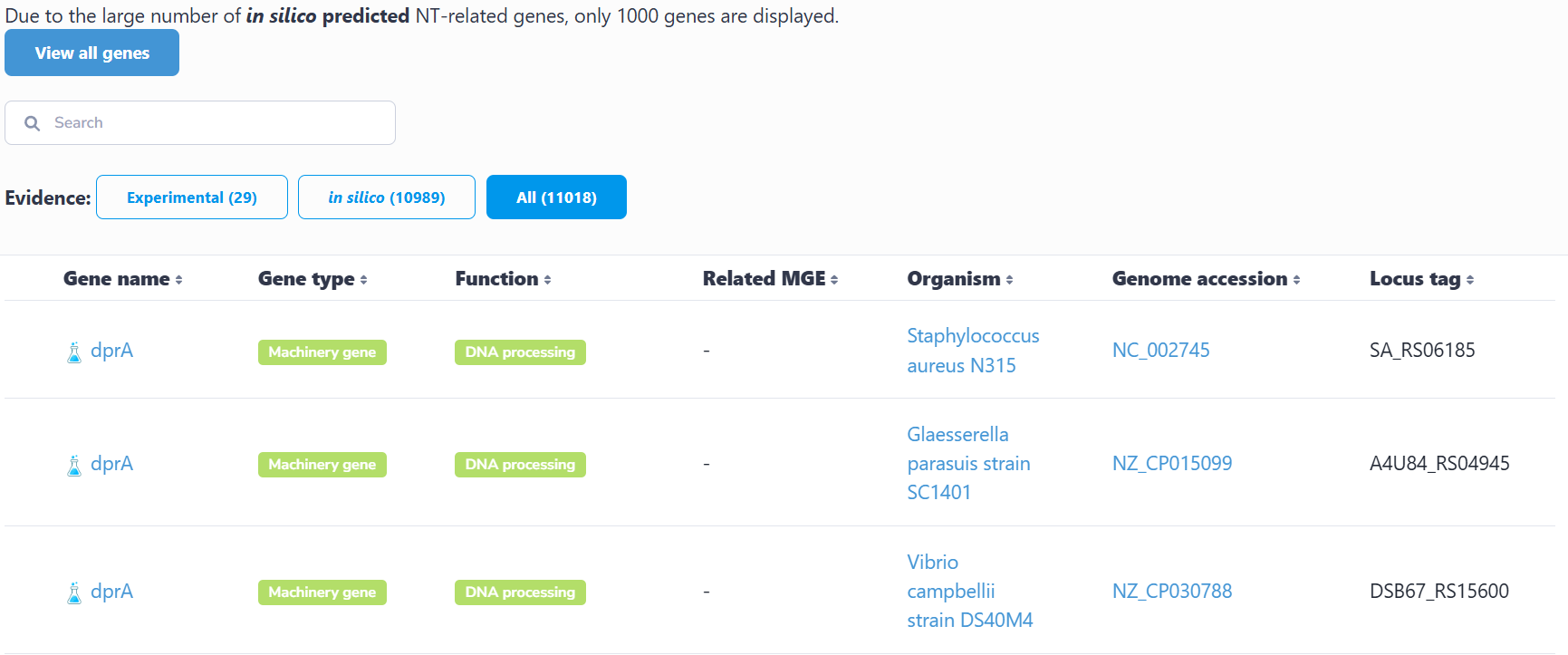
This interface catalogs all species containing experimentally verified or computationally predicted natural transformation machinery and regulatory genes. Users can navigate alphabetically or search via text input. Selection of a species accesses its strain list; subsequent strain selection displays associated genes. Clicking any gene retrieves its detailed information.
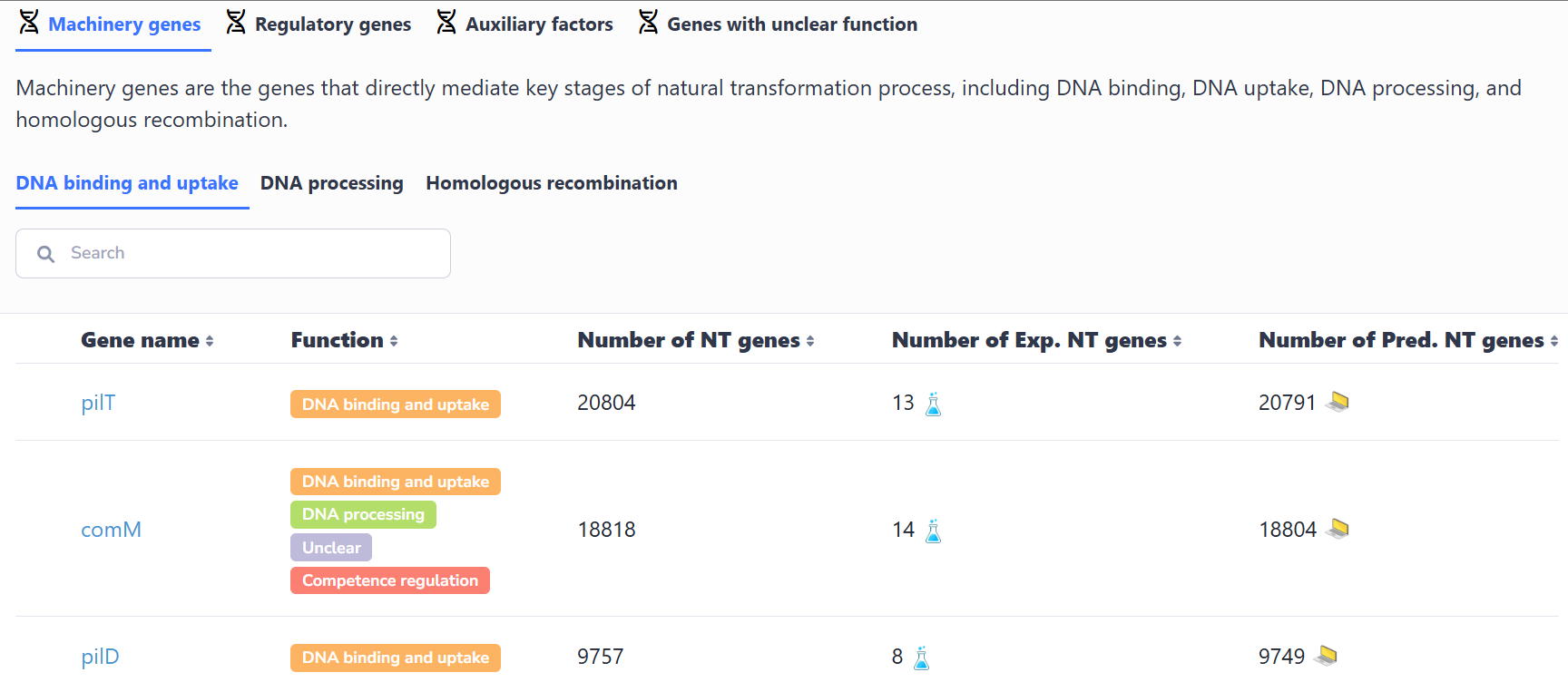
(2) Browse by function
Genes are categorized by functional roles within the natural transformation process (e.g., competence regulation, DNA binding, DNA uptake, homologous recombination). Selecting a functional category reveals relevant gene lists. Gene selection then displays taxonomic distribution across species, comprehensive gene list (experimental/predicted) and access to detailed gene records through individual selection.
On the "Statistics" page
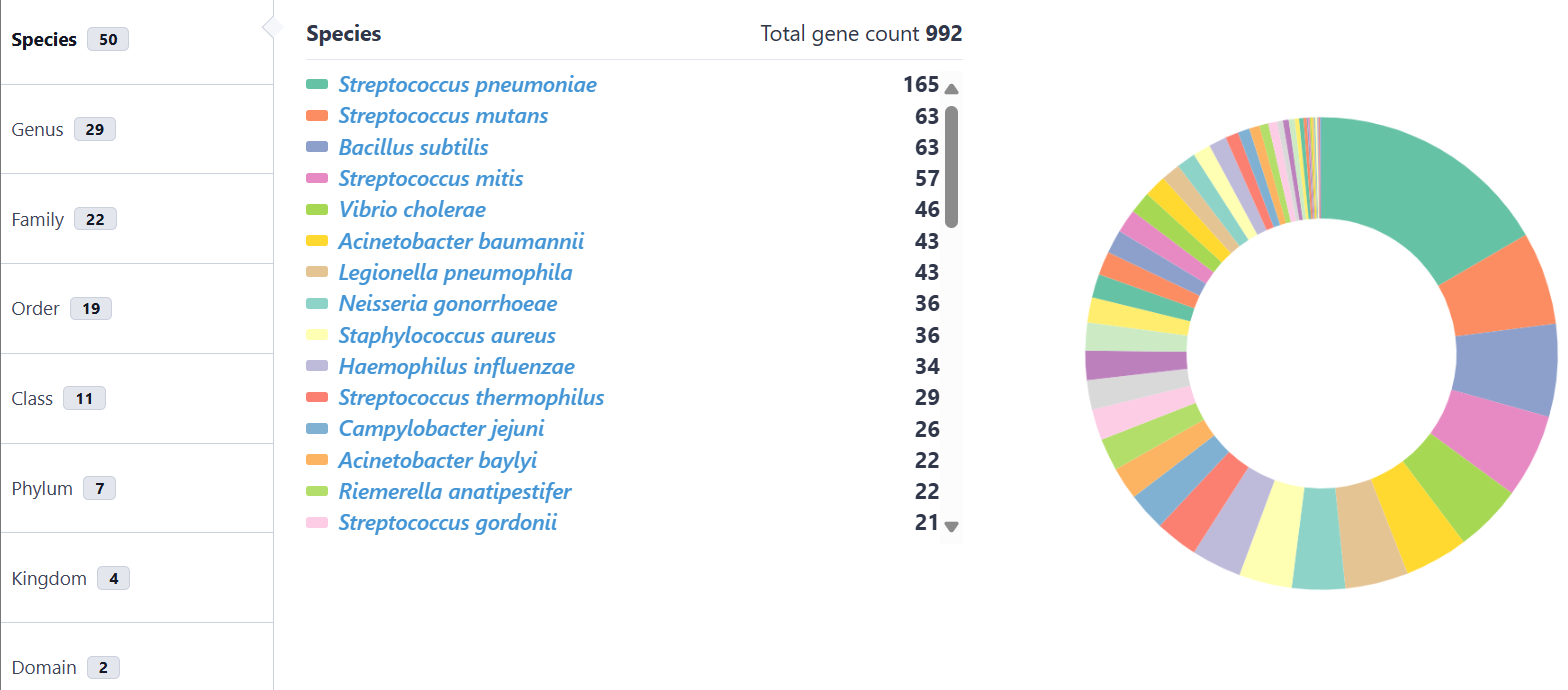
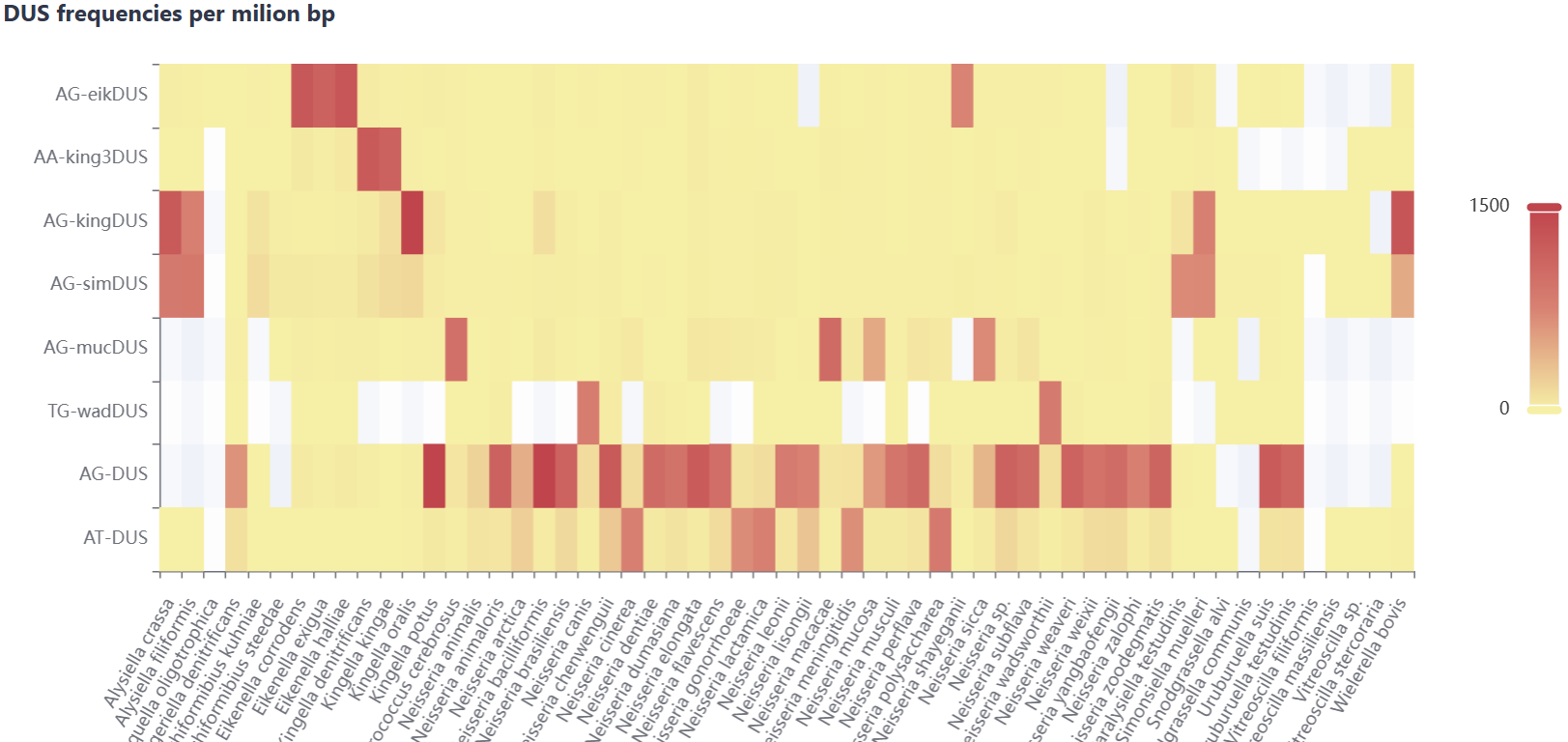
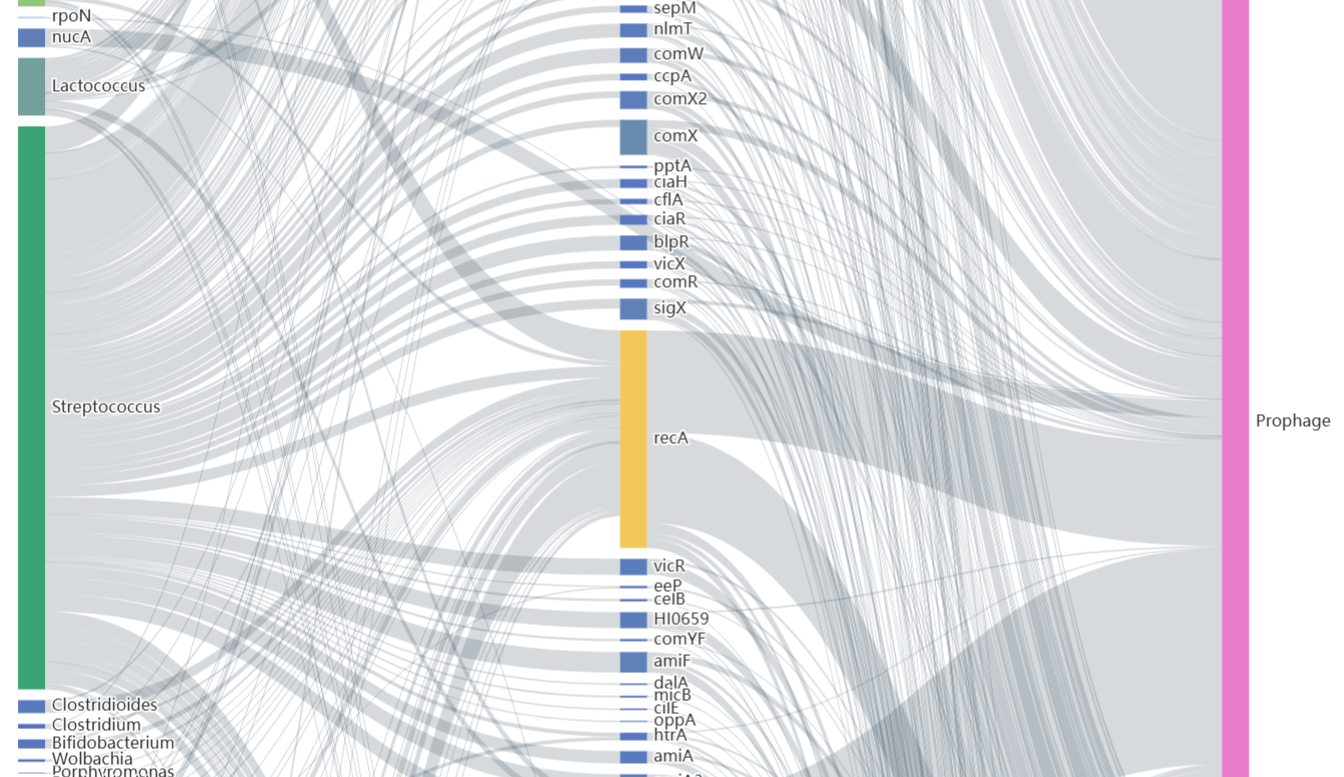
This interface dynamically visualizes the taxonomic distribution of the experimentally validated and in silico predicted genes within NTDB via interactive pie charts. Selection of specific taxa within these charts provides direct access to corresponding gene records. Additionally, natural transformable species without characterized genes are listed in a table, distributions of DNA uptake motifs (DUS/USS) rendered as interactive heatmaps, while MGE-gene-organism associations are represented through Sankey diagrams.
On the "Tools" page
In this interface, NTDB offers 4 online tools: Genome prediction, BLAST search, HMMER search and DUS/USS search.
On the "Download" page
Users can download the key datasets archived in NTDB on this page.
On the "References" page
This interface includes all the references related to natural transformation machinery and regulatory genes collected by NTDB, including some reviews. NTDB also supports the search of included references using keywords such as author, article title, journal, year, and PubMed ID.
FAQs
Q1. How to use NTDB?
A1: You can quickly understand the core functions of NTDB through the browse interface, and get more detailed through this Help manual.
Q2. In the entries collected by NTDB, some content is displayed as "-", what does this mean?
A2: If "-" appears in an experimentally validated entry, it indicates that we were unable to find corresponding data in the reference literature; if "-" appears in an in silico predicted entry, it means that the corresponding content was not predicted.
Q3: How could I contact you if I find an error or have suggestions for NTDB?
A3: Please do not hesitate to contact us through the mailing of hyou@sjtu.edu.cn.
Q4. Can I submit data to NTDB?
A4: You can contact Prof. Ou for cooperation through the mailing of hyou@sjtu.edu.cn.