
| 194_IME | |
| NBU2 | |
| - | |
| Bacteroides fragilis | |
| 11123 bp | |
| 3′ of two tRNASer genes | |
| linA2N2 (lincosamide resistance) | |
| - | |
| AF251288 (complete IME sequence in this GenBank file) | |
| - | |
| 1..11123 | |
| coordinates: 8812..9030; oriTDB id: 200011 TCCGCATGGAATACGGCAGAGAGCGGTACATCCGGGACGCTTCGCAGATTTACAGAGGATGCAAGGACTT GAACGAATACTTACAGAAACAGATTGAAAGAAAAAGGCAGCTCCAGTCCGCCAAAGGGGTGCGCAGCCAA TCACCGGAAAAGAAGAACGGCTTCCAGTTATAGCCCACTATAACTACACGCTTCGCTTTCGTAGTTGTGG GCTCTCCCG | |
| coordinates: 9137..10543; Gene: mobN2; Family: - |
The Interaction Network among ICE/IME/CIME/plasmid | ||||||
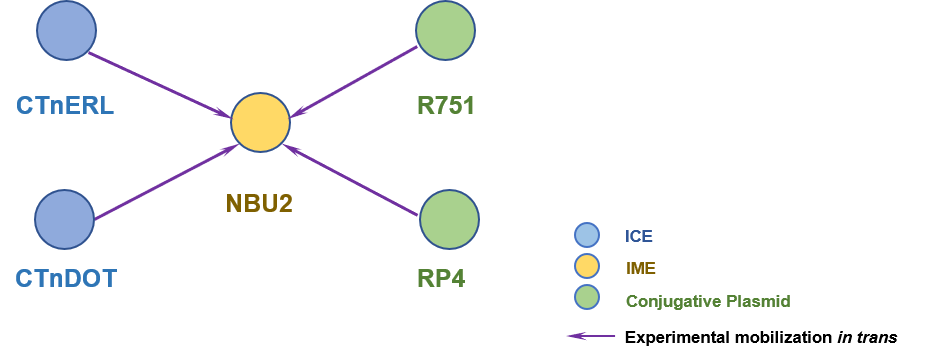 | ||||||
| Detailed Informatioin of the Interaction Network | ||||||
| # | IME | Inter_Ele | Methods | Donors | Recipients | Exper_Ref |
| 1 | NBU2 | CTnERL [ICE] | | Bacteroides fragilis; Bacteroides uniformis; Bacteroides thetaiotaomicron | Bacteroides thetaiotaomicron 5482; Bacteroides uniformis; Escherichia coli | 8407835; 7608064; 10852890 |
| 2 | NBU2 | RP4 [IncP plasmid] | | Escherichia coli | E. coli HB101 | 7608064 |
| 3 | NBU2 | R751 [IncP plasmid] | | Escherichia coli | E. coli HB101 | 7608064 |
| 4 | NBU2 | CTnDOT [ICE] | | Bacteroides thetaiotaomicron; Bacteroides uniformis | Bacteroides thetaiotaomicron 5482 | 8407835 |