The graph information of MGIVchMoz6 components from KC117175 |
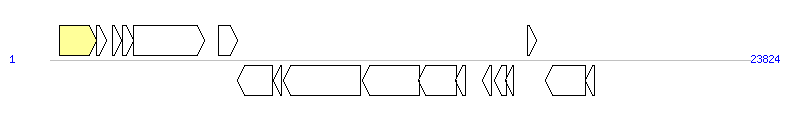 |
| Incomplete gene list of MGIVchMoz6 from KC117175 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | int | 340..1590 [+], 1251 | integrase family tyrosine site-specific recombinase | Integrase |
| 2 | MGIVCHMOZ6_002 | 1592..1930 [+], 339 | conserved hypothetical protein | |
| 3 | MGIVCHMOZ6_003 | 2126..2428 [+], 303 | conserved hypothetical protein | |
| 4 | MGIVCHMOZ6_004 | 2460..2831 [+], 372 | helix-turn-helix domain-containing protein | |
| 5 | MGIVCHMOZ6_005 | 2857..5265 [+], 2409 | DEAD/DEAH box helicase | |
| 6 | MGIVCHMOZ6_006 | 5723..6382 [+], 660 | conserved hypothetical protein | |
| 7 | MGIVCHMOZ6_007 | 6379..7581 [-], 1203 | hypothetical protein | |
| 8 | MGIVCHMOZ6_008 | 7591..7875 [-], 285 | hypothetical protein | |
| 9 | MGIVCHMOZ6_009 | 7953..10559 [-], 2607 | hypothetical protein | |
| 10 | MGIVCHMOZ6_010 | 10645..12570 [-], 1926 | hypothetical protein | |
| 11 | MGIVCHMOZ6_011 | 12557..13819 [-], 1263 | type II restriction-modification system methylation subunit | |
| 12 | MGIVCHMOZ6_012 | 13809..14141 [-], 333 | putative transcriptional regulator | |
| 13 | MGIVCHMOZ6_013 | 14705..15015 [-], 311 | hypothetical protein | |
| 14 | MGIVCHMOZ6_014 | 15116..15531 [-], 416 | hypothetical protein | |
| 15 | rdfM | 15518..15760 [-], 243 | recombination directionality factor | |
| 16 | MGIVCHMOZ6_016 | 16239..16553 [+], 315 | helix-turn-helix domain transcriptional regulator | |
| 17 | MGIVCHMOZ6_017 | 16873..18222 [-], 1350 | HipA domain-containing protein | |
| 18 | MGIVCHMOZ6_018 | 18212..18535 [-], 324 | helix-turn-helix domain transcriptional regulator | |

