The graph information of MGIVchHai6 components from AXDR01000001 |
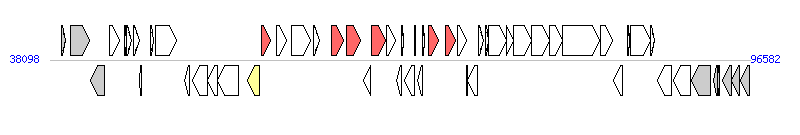 |
| Complete gene list of MGIVchHai6 from AXDR01000001 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | VCHC36A1_0040 | 39040..39375 [+], 336 | DNA binding, excisionase family domain protein | |
| 2 | VCHC36A1_0041 | 39818..41401 [+], 1584 | hypothetical protein | |
| 3 | VCHC36A1_0042 | 41455..42630 [-], 1176 | phage integrase family protein | |
| 4 | VCHC36A1_0043 | 43098..43940 [+], 843 | hypothetical protein | |
| 5 | VCHC36A1_0044 | 44355..44516 [+], 162 | hypothetical protein | |
| 6 | VCHC36A1_0045 | 44530..44943 [+], 414 | hypothetical protein | |
| 7 | VCHC36A1_0046 | 45077..45559 [+], 483 | resolvase, N terminal domain protein | |
| 8 | merR | 45587..45724 [-], 138 | mercuric resistance operon regulatory protein | |
| 9 | merP | 46487..46765 [+], 279 | mercuric transport protein periplasmic component | |
| 10 | merA | 46952..48661 [+], 1710 | mercuric reductase | |
| 11 | VCHC36A1_0050 | 49330..49755 [-], 426 | resolvase, N terminal domain protein | |
| 12 | VCHC36A1_0051 | 50005..51222 [-], 1218 | tniQ family protein | |
| 13 | VCHC36A1_0052 | 51219..52073 [-], 855 | bacterial TniB family protein | |
| 14 | VCHC36A1_0053 | 52130..53812 [-], 1683 | integrase core domain protein | |
| 15 | VCHC36A1_0054 | 54590..55603 [-], 1014 | integron integrase family protein | Integrase |
| 16 | VCHC36A1_0055 | 55749..56540 [+], 792 | nucleotidyltransferase domain protein | AR |
| 17 | VCHC36A1_0056 | 57045..57884 [+], 840 | dihydropteroate synthase | |
| 18 | VCHC36A1_0057 | 58289..59830 [+], 1542 | transposase family protein | |
| 19 | VCHC36A1_0058 | 60096..60593 [+], 498 | hypothetical protein | |
| 20 | VCHC36A1_0059 | 61651..62622 [+], 972 | dihydropteroate synthase | AR |
| 21 | VCHC36A1_0060 | 62839..64053 [+], 1215 | drug resistance transporter, Bcr/CflA subfamily protein | AR |
| 22 | tetR | 64260..64886 [-], 627 | tetracycline repressor protein class D | |
| 23 | tetA | 64990..66165 [+], 1176 | tetracycline resistance protein, class C | AR |
| 24 | VCHC36A1_0063 | 66186..66977 [+], 792 | bacterial regulatory helix-turn-helix, lysR family protein | |
| 25 | VCHC36A1_0064 | 67069..67476 [-], 408 | transposase | |
| 26 | VCHC36A1_0065 | 67492..67656 [+], 165 | putative transposase | |
| 27 | VCHC36A1_0066 | 67700..68563 [-], 864 | putative transposase | |
| 28 | VCHC36A1_0067 | 68515..68673 [+], 159 | hypothetical protein | |
| 29 | VCHC36A1_0068 | 68828..69226 [-], 399 | 60 kDa chaperonin domain protein | |
| 30 | intI1 | 69245..69391 [+], 147 | class 1 integrase domain protein | |
| 31 | pse4 | 69687..70553 [+], 867 | beta-lactamase PSE-4 | AR |
| 32 | VCHC36A1_0071 | 71111..71950 [+], 840 | dihydropteroate synthase | AR |
| 33 | VCHC36A1_0072 | 72164..72889 [+], 726 | hypothetical protein | |
| 34 | VCHC36A1_0073 | 72884..72997 [-], 114 | NTP-binding protein | |
| 35 | VCHC36A1_0074 | 73010..73831 [-], 822 | transposase | |
| 36 | VCHC36A1_0075 | 73891..74295 [+], 405 | resolvease/recombinase | |
| 37 | VCHC36A1_0076 | 74484..74714 [+], 231 | transcriptional regulator | |
| 38 | VCHC36A1_0077 | 74711..76210 [+], 1500 | N-6 DNA Methylase family protein | |
| 39 | VCHC36A1_0078 | 76207..76836 [+], 630 | hypothetical protein | |
| 40 | VCHC36A1_0079 | 76829..78295 [+], 1467 | divergent AAA domain protein | |
| 41 | VCHC36A1_0080 | 78292..79794 [+], 1503 | N-6 DNA Methylase family protein | |
| 42 | VCHC36A1_0081 | 79795..80934 [+], 1140 | type I restriction modification DNA specificity domain protein | |
| 43 | hsdR | 80940..84002 [+], 3063 | type I site-specific deoxyribonuclease, HsdR family protein | |
| 44 | VCHC36A1_0083 | 84053..85087 [+], 1035 | putative DNA-binding protein | |
| 45 | VCHC36A1_0084 | 85145..85939 [-], 795 | hypothetical protein | Relaxase |
| 46 | VCHC36A1_0085 | 86351..86548 [+], 198 | DNA binding, excisionase family domain protein | |
| 47 | VCHC36A1_0086 | 86561..88237 [+], 1677 | hypothetical protein | |
| 48 | VCHC36A1_0087 | 88299..88643 [+], 345 | DNA binding, excisionase family domain protein | |
| 49 | VCHC36A1_0088 | 88888..90057 [-], 1170 | phage integrase family protein | |
| 50 | trmE | 90221..91582 [-], 1362 | tRNA modification GTPase TrmE | |
| 51 | oxaA | 91676..93301 [-], 1626 | inner membrane protein oxaA | |
| 52 | rnpA | 93528..93851 [-], 324 | ribonuclease P protein component | |
| 53 | rpmH | 93897..94034 [-], 138 | ribosomal protein L34 | |
| 54 | VCHC36A1_0093 | 94276..95013 [-], 738 | ABC transporter family protein | |
| 55 | yxeN | 95010..95681 [-], 672 | putative amino-acid permease protein yxeN | |
| 56 | yxeM | 95759..96505 [-], 747 | putative amino-acid-binding protein yxeM | |