
| 961 | |
| ICEApl2 | |
| ICEO_0000144 | |
| Actinobacillus pleuropneumoniae serovar 8 | |
| 92660 | |
| 46.94 | |
| prfC | |
| Antibiotic resistance genes: floR, strAB, sul2 and dfrA1 | |
| Actinobacillus pleuropneumoniae | |
| MF187965 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..92660 | |
| coordinates: 6516..6622; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 38193..40343; Locus tag: ICEApl2.36; Family: MOBH |
The graph information of ICEApl2 components from MF187965 | |||||
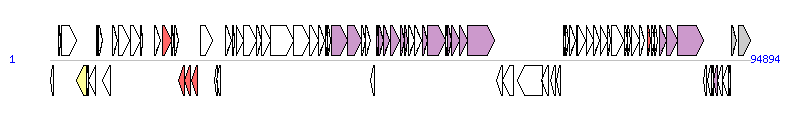 | |||||
| Complete gene list of ICEApl2 from MF187965 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | yhaH | 3..470 [-], 468 | YhaH | ||
| 2 | ICEApl2.01 | 1175..1363 [+], 189 | DNA-binding protein | ||
| 3 | ICEApl2.02 | 1590..3527 [+], 1938 | hypothetical protein | ||
| 4 | intS | 3657..4898 [-], 1242 | hypothetical protein | Integrase | |
| 5 | ICEApl2.04 | 4900..5169 [-], 270 | hypothetical protein | ||
| 6 | ICEApl2.05 | 5172..6146 [-], 975 | rod shape determination protein | ||
| 7 | ICEApl2.06 | 6242..6409 [+], 168 | hypothetical protein | ||
| 8 | ICEApl2.07 | 6611..7054 [+], 444 | hypothetical protein | ||
| 9 | ICEApl2.08 | 7065..8237 [-], 1173 | hypothetical protein | ||
| 10 | ICEApl2.09 | 8521..9114 [+], 594 | hypothetical protein | ||
| 11 | tnp | 9228..10724 [+], 1497 | hypothetical protein | ||
| 12 | ICEApl2.11 | 10866..12206 [+], 1341 | hypothetical protein | ||
| 13 | ICEApl2.12 | 12321..12512 [+], 192 | RepA protein | ||
| 14 | ICEApl2.14 | 14147..15031 [+], 885 | T4SS protein | ||
| 15 | floR | 15248..16462 [+], 1215 | hypothetical protein | AR | |
| 16 | ICEApl2.16 | 16490..16795 [+], 306 | putative transcriptional regulator | ||
| 17 | tnpA | 16907..17446 [+], 540 | hypothetical protein | ||
| 18 | strB | 17418..18254 [-], 837 | hypothetical protein | AR | |
| 19 | strA | 18254..19057 [-], 804 | hypothetical protein | AR | |
| 20 | sul2 | 19118..19933 [-], 816 | hypothetical protein | AR | |
| 21 | ICEApl2.21 | 20404..22017 [+], 1614 | transposase Tn3 family protein | ||
| 22 | ICEApl2.22 | 22360..22668 [-], 309 | DNA polymerase V subunit UmuC | ||
| 23 | lexA_1 | 22637..23086 [-], 450 | LexA repressor | ||
| 24 | polC | 23725..24630 [+], 906 | PolC-type DNA polymerase III | ||
| 25 | ICEApl2.25 | 24718..25017 [+], 300 | hypothetical protein | ||
| 26 | ICEApl2.26 | 25339..26289 [+], 951 | hypothetical protein | ||
| 27 | hsdM | 26298..28013 [+], 1716 | type I restriction enzyme EcoKI M protein | ||
| 28 | ICEApl2.28 | 28006..28710 [+], 705 | hypothetical protein | ||
| 29 | ICEApl2.29 | 28707..29861 [+], 1155 | BstXI restriction endonuclease | ||
| 30 | hsdR_1 | 29866..33030 [+], 3165 | type I restriction enzyme EcoR124II R protein | ||
| 31 | mcrB | 33046..35244 [+], 2199 | 5-methylcytosine-specific restriction enzyme B | ||
| 32 | ICEApl2.32 | 35244..36371 [+], 1128 | hypothetical protein | ||
| 33 | mrr | 36371..37225 [+], 855 | Mrr restriction system protein | ||
| 34 | ICEApl2.34 | 37319..37600 [+], 282 | hypothetical protein | ||
| 35 | ICEApl2.35 | 37600..38073 [+], 474 | hypothetical protein | ||
| 36 | ICEApl2.36 | 38193..40343 [+], 2151 | putative helicase | Relaxase, MOBH Family | |
| 37 | ICEApl2.37 | 40392..42212 [+], 1821 | AAA-like domain protein | TraD_F, T4SS component | |
| 38 | ICEApl2.38 | 42222..42782 [+], 561 | hypothetical protein | ||
| 39 | ICEApl2.39 | 42769..43404 [+], 636 | hypothetical protein | ||
| 40 | ICEApl2.40 | 43431..44018 [-], 588 | replicative DNA helicase | ||
| 41 | ICEApl2.41 | 44307..44588 [+], 282 | TraL protein | TraL_F, T4SS component | |
| 42 | ICEApl2.42 | 44585..45211 [+], 627 | TraE protein | TraE_F, T4SS component | |
| 43 | ICEApl2.43 | 45192..46091 [+], 900 | TraK protein | TraK_F, T4SS component | |
| 44 | ICEApl2.44 | 46094..47383 [+], 1290 | bacterial conjugation TrbI-like protein | TraB_F, T4SS component | |
| 45 | ICEApl2.45 | 47458..48030 [+], 573 | hypothetical protein | TraV_F, T4SS component | |
| 46 | ICEApl2.46 | 48027..48413 [+], 387 | hypothetical protein | TraA_F, T4SS component | |
| 47 | ICEApl2.47 | 48592..49425 [+], 834 | hypothetical protein | ||
| 48 | ICEApl2.48 | 49418..50356 [+], 939 | hypothetical protein | ||
| 49 | dsbC_1 | 50488..51180 [+], 693 | thiol:disulfide interchange protein DsbC | TrbB_I, T4SS component | |
| 50 | ICEApl2.50 | 51180..53579 [+], 2400 | AAA-like domain protein | TraC_F, T4SS component | |
| 51 | ICEApl2.51 | 53572..53919 [+], 348 | hypothetical protein | ||
| 52 | ICEApl2.52 | 53903..54415 [+], 513 | peptidase S26 | TraF, T4SS component | |
| 53 | ICEApl2.53 | 54426..55550 [+], 1125 | hypothetical protein | TraW_F, T4SS component | |
| 54 | ICEApl2.54 | 55534..56562 [+], 1029 | TraU protein | TraU_F, T4SS component | |
| 55 | ICEApl2.55 | 56565..60257 [+], 3693 | hypothetical protein | TraN_F, T4SS component | |
| 56 | dnaI | 60594..61349 [-], 756 | primosomal protein DnaI | ||
| 57 | ICEApl2.57 | 61361..62881 [-], 1521 | integrase core domain protein | ||
| 58 | ICEApl2.58 | 63333..66698 [-], 3366 | ski2-like helicase | ||
| 59 | ICEApl2.59 | 66685..67560 [-], 876 | hypothetical protein | ||
| 60 | endA | 67832..68515 [-], 684 | endonuclease-1 precursor | ||
| 61 | ICEApl2.61 | 68624..69226 [-], 603 | hypothetical protein | ||
| 62 | ICEApl2.62 | 69596..69922 [+], 327 | hypothetical protein | ||
| 63 | ssb_2 | 69938..70357 [+], 420 | single-stranded DNA-binding protein | ||
| 64 | ICEApl2.64 | 70437..71255 [+], 819 | RecT family | ||
| 65 | ICEApl2.65 | 71339..71482 [+], 144 | hypothetical protein | ||
| 66 | ICEApl2.66 | 71543..72559 [+], 1017 | YqaJ-like viral recombinase domain | ||
| 67 | cobS | 72769..73728 [+], 960 | aerobic cobaltochelatase subunit CobS | ||
| 68 | ICEApl2.68 | 73728..74495 [+], 768 | hypothetical protein | ||
| 69 | ICEApl2.69 | 74594..75547 [+], 954 | hypothetical protein | ||
| 70 | ICEApl2.70 | 75609..76049 [+], 441 | hypothetical protein | ||
| 71 | ICEApl2.71 | 76119..77774 [+], 1656 | cobalamin biosynthesis protein CobT | ||
| 72 | ICEApl2.72 | 77858..78355 [+], 498 | hypothetical protein | ||
| 73 | ICEApl2.73 | 78355..78696 [+], 342 | hypothetical protein | ||
| 74 | ICEApl2.74 | 78788..79861 [+], 1074 | primase-helicase zinc-binding domain | ||
| 75 | ICEApl2.75 | 79951..80658 [+], 708 | hypothetical protein | ||
| 76 | dhfrI | 80964..81437 [+], 474 | dihydrofolate reductase type 1 | AR | |
| 77 | ICEApl2.77 | 81550..81912 [+], 363 | hypothetical protein | ||
| 78 | ICEApl2.78 | 82007..82420 [+], 414 | hypothetical protein | ||
| 79 | ICEApl2.79 | 82667..83611 [+], 945 | hypothetical protein | TraF_F, T4SS component | |
| 80 | ICEApl2.80 | 83614..85002 [+], 1389 | hypothetical protein | TraH_F, T4SS component | |
| 81 | ICEApl2.81 | 85006..88575 [+], 3570 | TraG-like protein | TraG_F, T4SS component | |
| 82 | ICEApl2.82 | 88608..89039 [-], 432 | hypothetical protein | ||
| 83 | flhC | 89097..89630 [-], 534 | flagellar transcriptional regulator FlhC | ||
| 84 | ICEApl2.84 | 89627..89926 [-], 300 | transcriptional activator FlhD | ||
| 85 | ICEApl2.85 | 89923..90471 [-], 549 | transglycosylase SLT domain protein | Orf169_F, T4SS component | |
| 86 | ICEApl2.86 | 90458..91120 [-], 663 | hypothetical protein | ||
| 87 | ICEApl2.87 | 91107..91976 [-], 870 | hypothetical protein | ||
| 88 | ICEApl2.88 | 92032..92283 [-], 252 | hypothetical protein | ||
| 89 | ICEApl2.89 | 92401..93048 [+], 648 | hypothetical protein | ||
| 90 | prfC | 93305..94894 [+], 1590 | PrfC | ||
| (1) Li Y; Li Y; Fernandez Crespo R; Leanse LG; Langford PR; Bosse JT (2018). Characterization of the Actinobacillus pleuropneumoniae SXT-related integrative and conjugative element ICEApl2 and analysis of the encoded FloR protein: hydrophobic residues in transmembrane domains contribute dynamically to florfenicol and chloramphenicol efflux. J Antimicrob Chemother. 73(1):57-65. [PubMed:29029160] |