
| 80 | |
| ICESpn8140 | |
| Streptococcus pneumoniae 8140 | |
| 26888 | |
| 36.06 | |
| - | |
| - | |
| - | |
| FR671412 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..26888 | |
| coordinates: 15331..15543; oriTDB id: 200004 GCCTGACCGCAGGTCAGGCGCAGACCGTAGCCCGAAGTCTCTAGGCCATGATGAAACTTCAACCCCCGTT TTCTAATAGGGGGGTTACATTTGGCAAATACGCCAAGTCCACCCCTCCTCATTCCTTATGGGAGTTGGGT TTTCAATTTTTATGAAGTGTGTCACTTTCGTCAAAAAAGTGTGTCACTTTGTTAGAAAGAGGTGTTGCGA TGA | |
| coordinates: 15544..16776; Family: MOBT |
The graph information of ICESpn8140 components from FR671412 | |||||
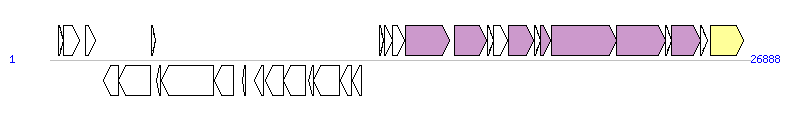 | |||||
| Complete gene list of ICESpn8140 from FR671412 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 323..511 [+], 189 | Conserved hypothetical protein | ||
| 2 | - | 504..1115 [+], 612 | Fic/DOC domain protein | ||
| 3 | - | 1370..1732 [+], 363 | Membrane protein | ||
| 4 | - | 2050..2646 [-], 597 | Response regulator | ||
| 5 | - | 2627..3868 [-], 1242 | Sensor histidine kinase | ||
| 6 | - | 3890..4060 [+], 171 | Hypothetical protein | ||
| 7 | - | 4104..4244 [-], 141 | Hypothetical protein | ||
| 8 | - | 4282..6297 [-], 2016 | ABC transporter, permease subunit | ||
| 9 | - | 6310..7047 [-], 738 | ABC-type antimicrobial peptide transport system, ATPase component | ||
| 10 | - | 7407..7520 [-], 114 | Hypothetical protein | ||
| 11 | - | 7851..8213 [-], 363 | Hypothetical protein | ||
| 12 | - | 8270..8956 [-], 687 | Helix-turn-helix DNA binding protein | ||
| 13 | - | 8965..9825 [-], 861 | Conserved hypothetical protein | ||
| 14 | - | 9946..10131 [-], 186 | Conserved hypothetical protein | ||
| 15 | - | 10112..11125 [-], 1014 | Conserved hypothetical protein | ||
| 16 | - | 11155..11571 [-], 417 | Putative FMN-binding protein | ||
| 17 | - | 11626..11976 [-], 351 | Helix-turn-helix DNA binding protein | ||
| 18 | - | 12638..12820 [+], 183 | Hypothetical protein | ||
| 19 | - | 12855..13151 [+], 297 | Conserved hypothetical protein | ||
| 20 | - | 13173..13634 [+], 462 | Conserved hypothetical protein | ||
| 21 | - | 13648..15336 [+], 1689 | FtsK/SpoIIIE protein | YdcQ, T4SS component | |
| 22 | - | 15544..16776 [+], 1233 | Helix-turn-helix DNA binding protein | Relaxase, MOBT Family | |
| 23 | - | 16791..17024 [+], 234 | Conserved hypothetical protein | ||
| 24 | - | 17026..17583 [+], 558 | Putative transfer protein | ||
| 25 | - | 17594..18589 [+], 996 | Putative transfer protein | YddB, T4SS component | |
| 26 | - | 18599..18823 [+], 225 | Conserved hypothetical protein | ||
| 27 | - | 18826..19236 [+], 411 | Putative transfer protein | YddD, T4SS component | |
| 28 | - | 19250..21754 [+], 2505 | Putative conjugative transfer ATP/GTP-binding protein | YddE, T4SS component | |
| 29 | - | 21766..23631 [+], 1866 | Putative conjugative transfer membrane-associated protein | YddG, T4SS component | |
| 30 | - | 23633..23857 [+], 225 | Putative conjugative transfer protein | ||
| 31 | - | 23874..24986 [+], 1113 | Cysteine/histidine-dependent peptidase | Orf13_p, T4SS component | |
| 32 | - | 25000..25257 [+], 258 | Putative DNA binding conjugative transfer protein | ||
| 33 | - | 25369..26634 [+], 1266 | Integrase | Integrase | |
|
(1) Croucher NJ; Harris SR; Fraser C; Quail MA; Burton J; van der Linden M; McGee L; von Gottberg A; Song JH; Ko KS; Pichon B; Baker S; Parry CM; Lambertsen LM; Shahinas D; Pillai DR; Mitchell TJ; Dougan G; Tomasz A; Klugman KP; Parkhill J; Hanage WP; Bentley SD (2011). Rapid pneumococcal evolution in response to clinical interventions. Science. 331(6016):430-4. [PubMed:21273480] |