
| 751 | |
| ICE_SsaN20_rpsI | |
| Streptococcus salivarius N20 | |
| 37,356 | |
| 34.46 | |
| rpsI | |
| - | |
| - | |
| LT622836 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..37356 | |
| coordinates: 24568..24780; oriTDB id: 200003 GCCTGACCACAGGTCAGGCGCAGACCGTAGCCCGAAGTTCCTAGGCCATGATGAGACTTCAACCCCCGAT TTCTAATAGGGGGGTTACATTTGGCCAAAGTGCCACGTCCACCCCTCCTCATTCCTTGTGGGAGTTGGGA TTTCAAGATTTAGAAAGTGTGTCACTTTGGTCCAAAAAGTGTGTCACTTAATTAAAAGGAGGTGTTTCCA TGA | |
| coordinates: 24781..26013; Gene: ORFJ; Family: MOBT |
The graph information of ICE_SsaN20_rpsI components from LT622836 | |||||
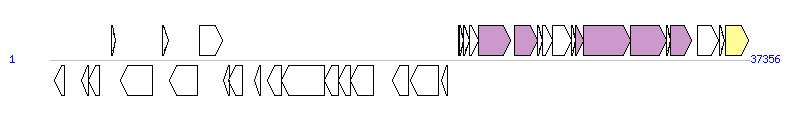 | |||||
| Complete gene list of ICE_SsaN20_rpsI from LT622836 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 217..798 [-], 582 | hypothetical protein | ||
| 2 | cadX | 1694..2032 [-], 339 | Cadmium efflux system accessory protein CadX | ||
| 3 | cadD | 2044..2658 [-], 615 | Cadmium resistance protein CadD | ||
| 4 | - | 3264..3482 [+], 219 | hypothetical protein | ||
| 5 | - | 3763..5484 [-], 1722 | hypothetical protein | ||
| 6 | - | 5995..6300 [+], 306 | hypothetical protein | ||
| 7 | sth368IR | 6363..7847 [-], 1485 | R.Sth368I endonuclease | ||
| 8 | sth368IM | 7963..9228 [+], 1266 | Cytosine-specific methyltransferase | ||
| 9 | - | 9258..9557 [-], 300 | hypothetical protein | ||
| 10 | arp1 | 9550..10248 [-], 699 | putative transcriptional regulator Arp1 | ||
| 11 | tnp | 10919..11242 [-], 324 | Transposase | ||
| 12 | - | 11608..12357 [-], 750 | hypothetical protein | ||
| 13 | - | 12368..14656 [-], 2289 | hypothetical protein | ||
| 14 | - | 14659..15405 [-], 747 | hypothetical protein | ||
| 15 | - | 15402..16019 [-], 618 | hypothetical protein | ||
| 16 | - | 16023..17270 [-], 1248 | hypothetical protein | ||
| 17 | ORFQ | 18262..19122 [-], 861 | putative ImmA protease | ||
| 18 | - | 19215..20708 [-], 1494 | hypothetical protein | ||
| 19 | arp2 | 20881..21216 [-], 336 | putative transcriptional regulator Arp2 | ||
| 20 | ORFO | 21806..21835 [+], 30 | conserved protein of unknown function | ||
| 21 | ORFN | 21870..22052 [+], 183 | conserved protein of unknown function | ||
| 22 | ORFM | 22091..22387 [+], 297 | Putative conjugal transfer protein | ||
| 23 | ORFL | 22410..22871 [+], 462 | Putative conjugal transfer protein | ||
| 24 | ORFK | 22885..24573 [+], 1689 | FtsK/SpoIIIE family coupling protein | YdcQ, T4SS component | |
| 25 | ORFJ | 24781..26013 [+], 1233 | Putative relaxase | Relaxase, MOBT Family | |
| 26 | ORFI | 26027..26257 [+], 231 | Putative conjugal transfer protein | ||
| 27 | ORFH | 26261..26818 [+], 558 | Putative conjugal transfer protein | ||
| 28 | ORFG | 26830..27825 [+], 996 | Putative conjugal transfer protein | ||
| 29 | ORFF | 27835..28059 [+], 225 | Putative conjugal transfer protein | ||
| 30 | ORFE | 28062..28472 [+], 411 | Putative conjugal transfer protein | YddD, T4SS component | |
| 31 | ORFD | 28486..30990 [+], 2505 | Putative conjugal transfer protein | YddE, T4SS component | |
| 32 | ORFC | 31002..32882 [+], 1881 | Putative conjugal transfer protein | YddG, T4SS component | |
| 33 | ORFB | 32884..33108 [+], 225 | Putative conjugal transfer protein | ||
| 34 | ORFA | 33125..34237 [+], 1113 | Putative conjugal transfer protein | Orf13_p, T4SS component | |
| 35 | - | 34550..35692 [+], 1143 | hypothetical protein | ||
| 36 | xis | 35747..36019 [+], 273 | Excisionase | ||
| 37 | int | 36030..37250 [+], 1221 | Tyrosine integrase | Integrase | |
|
(1) Dahmane N; Libante V; Charron-Bourgoin F; Guedon E; Guedon G; Leblond-Bourget N; Payot S (2017). Diversity of Integrative and Conjugative Elements of Streptococcus salivarius and Their Intra- and Interspecies Transfer. Appl Environ Microbiol. 83(13). [PubMed:28432093] |