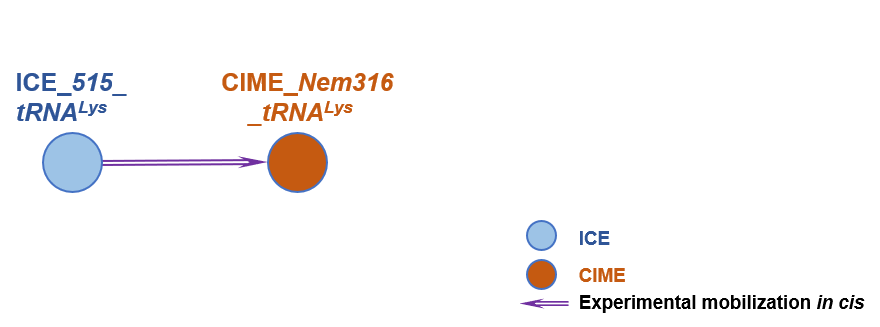
The graph information of ICE_515_tRNALys components from AAJP01000002 |
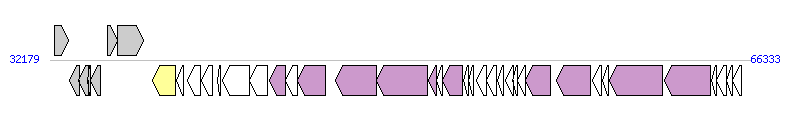 |
| Incomplete gene list of ICE_515_tRNALys from AAJP01000002 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | SAL_2022 | 32384..33100 [+], 717 | transcriptional regulator, MerR family | |
| 2 | SAL_2023 | 33139..33609 [-], 471 | acetyltransferase, GNAT family | |
| 3 | SAL_2024 | 33581..34039 [-], 459 | MutT/nudix family protein | |
| 4 | SAL_2025 | 34026..34169 [-], 144 | conserved hypothetical protein | |
| 5 | SAL_2026 | 34175..34645 [-], 471 | conserved hypothetical protein | |
| 6 | SAL_2027 | 34972..35463 [+], 492 | acetyltransferase, GNAT family | |
| 7 | SAL_2028 | 35456..36724 [+], 1269 | ATPase, AAA family | |
| 8 | SAL_2029 | 37179..38300 [-], 1122 | site-specific recombinase, phage integrase family | Integrase |
| 9 | SAL_2030 | 38350..38676 [-], 327 | Domain of unknown function (DUF771) superfamily | |
| 10 | SAL_2031 | 38869..39501 [-], 633 | conserved hypothetical protein | |
| 11 | SAL_2032 | 39577..40119 [-], 543 | Unknown | |
| 12 | SAL_2033 | 40342..40503 [-], 162 | putative cell wall surface anchor family protein | |
| 13 | SAL_2034 | 40605..41921 [-], 1317 | conserved hypothetical protein | |
| 14 | SAL_2035 | 41923..42792 [-], 870 | putative DNA adenine methylase | |
| 15 | SAL_2036 | 42888..43667 [-], 780 | cell wall surface anchor signal protein | PrgC, T4SS component |
| 16 | SAL_2037 | 43684..44232 [-], 549 | conserved hypothetical protein | |
| 17 | SAL_2038 | 44232..45605 [-], 1374 | CHAP domain family | PrgK, T4SS component |
| 18 | SAL_2039 | 46101..48101 [-], 2001 | putative membrane protein | YddG, T4SS component |
| 19 | SAL_2040 | 48117..50618 [-], 2502 | conjugal transfer protein, interruption-C | YddE, T4SS component |
| 20 | SAL_2041 | 50639..51013 [-], 375 | conserved hypothetical protein | YddD, T4SS component |
| 21 | SAL_2042 | 51053..51319 [-], 267 | conserved hypothetical protein | |
| 22 | SAL_2043 | 51340..52293 [-], 954 | conserved hypothetical protein | YddB, T4SS component |
| 23 | SAL_2044 | 52346..52600 [-], 255 | conserved hypothetical protein | |
| 24 | SAL_2045 | 52593..52859 [-], 267 | Uncharacterized ACR, COG2161 subfamily | |
| 25 | SAL_2046 | 53004..53477 [-], 474 | conserved hypothetical protein | |
| 26 | SAL_2047 | 53481..53966 [-], 486 | conserved hypothetical protein | |
| 27 | SAL_2048 | 54019..54291 [-], 273 | conserved hypothetical protein | |
| 28 | SAL_2049 | 54423..54776 [-], 354 | transcriptional regulator, Cro/CI family | |
| 29 | SAL_2050 | 54769..54999 [-], 231 | transcriptional regulator, Cro/CI family | |
| 30 | SAL_2051 | 54986..55363 [-], 378 | conserved hypothetical protein | |
| 31 | SAL_2052 | 55379..56611 [-], 1233 | transcriptional regulator, Cro/CI family | Relaxase, MOBT Family |
| 32 | SAL_2053 | 56899..58560 [-], 1662 | FtsK/SpoIIIE family protein | YdcQ, T4SS component |
| 33 | SAL_2054 | 58637..59098 [-], 462 | conserved hypothetical protein | |
| 34 | SAL_2055 | 59120..59416 [-], 297 | conserved hypothetical protein | |
| 35 | SAL_2056 | 59476..62067 [-], 2592 | cell wall surface anchor family protein | PrgB, T4SS component |
| 36 | SAL_2057 | 62156..64417 [-], 2262 | Unknown | PrgA, T4SS component |
| 37 | SAL_2058 | 64433..64696 [-], 264 | conserved hypothetical protein | |
| 38 | SAL_2059 | 64716..65177 [-], 462 | Unknown | |
| 39 | SAL_2060 | 65162..65473 [-], 312 | conserved hypothetical protein | |
| 40 | SAL_2061 | 65466..65897 [-], 432 | transcriptional regulator, Cro/CI family | |