The graph information of HPI-ICEEh1 components from FN297818 |
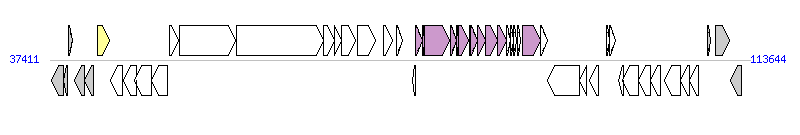 |
| Complete gene list of HPI-ICEEh1 from FN297818 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | - | 37601..38842 [-], 1242 | hypothetical protein | |
| 2 | - | 38861..39280 [-], 420 | hypothetical protein | |
| 3 | - | 39392..39895 [+], 504 | putative Fe2+/Zn2+ uptake regulation protein | |
| 4 | - | 40101..41120 [-], 1020 | hypothetical protein | |
| 5 | - | 41120..42106 [-], 987 | putative iron(III) ABC-transporter periplasmic-binding protein | |
| 6 | intB | 42589..43851 [+], 1263 | integrase | Integrase |
| 7 | ybtS | 44045..45349 [-], 1305 | salicylate synthetase | |
| 8 | ybtX | 45377..46780 [-], 1404 | cytoplasmic transmembrane protein | |
| 9 | ybtQ | 46650..48452 [-], 1803 | ABC-transporter protein | |
| 10 | ybtP | 48439..50241 [-], 1803 | inner membrane ABC-transporter | |
| 11 | ybtA | 50408..51367 [+], 960 | transcriptional regulator | |
| 12 | irp2 | 51558..57665 [+], 6108 | High Molecular Weight Protein 2 (HMWP2) | |
| 13 | irp1 | 57753..67244 [+], 9492 | High Molecular Weight Protein 1 (HMWP1) | |
| 14 | ybtU | 67145..68341 [+], 1197 | thiazolinyl-S-HMWP1 reductase | |
| 15 | ybtT | 68353..69141 [+], 789 | yersiniabactin biosynthetic protein | |
| 16 | ybtE | 69145..70722 [+], 1578 | yersiniabactin biosynthetic protein | |
| 17 | fyuA | 70853..72874 [+], 2022 | yersiniabactin/pesticin receptor | |
| 18 | - | 73747..74673 [+], 927 | hypothetical protein | |
| 19 | - | 75187..75768 [+], 582 | hypothetical protein | |
| 20 | - | 76905..77189 [-], 285 | hypothetical protein | |
| 21 | virB1-like2 | 77213..77923 [+], 711 | putative Pilx1/VirB1-like protein | VirB1, T4SS component |
| 22 | virB2-like2 | 77923..78216 [+], 294 | putative VirB2-like protein | VirB2, T4SS component |
| 23 | virB3-4-like2 | 78229..80967 [+], 2739 | putative Pilx3-4/VirB3-4-like protein | VirB4, T4SS component |
| 24 | virB5-like2 | 80985..81692 [+], 708 | putative Pilx5/VirB5-like protein | VirB5, T4SS component |
| 25 | - | 81700..81942 [+], 243 | hypothetical protein | MagB05, T4SS component |
| 26 | virB6-like2 | 81946..83019 [+], 1074 | putative Pilx6/VirB6-like protein | VirB6, T4SS component |
| 27 | - | 83111..83248 [+], 138 | hypothetical protein | VirB7, T4SS component |
| 28 | virB8-like2 | 83241..83924 [+], 684 | putative PilX8-VirB8-like protein | VirB8, T4SS component |
| 29 | virB9-like2 | 83921..84829 [+], 909 | putative Pilx9/VirB9-like protein | VirB9, T4SS component |
| 30 | virB10-like2 | 84873..86123 [+], 1251 | putative Pilx10/VirB10-like protein | VirB10, T4SS component |
| 31 | virB11-like2 | 86113..87138 [+], 1026 | putative Pilx11/VirB11-like protein | VirB11, T4SS component |
| 32 | - | 87135..87533 [+], 399 | hypothetical protein | |
| 33 | - | 87569..87874 [+], 306 | YggA-like protein | |
| 34 | - | 87908..88213 [+], 306 | hypothetical protein | |
| 35 | - | 88324..88626 [+], 303 | hypothetical protein | |
| 36 | mobB-like2 | 88893..90782 [+], 1890 | putative MobB-like protein | VirD4, T4SS component |
| 37 | mobC-like2 | 90792..91538 [+], 747 | MobC-like protein | |
| 38 | - | 91599..95123 [-], 3525 | helicase-like protein | |
| 39 | vrlR-like2 | 95107..95886 [-], 780 | putative VrlR-like protein | |
| 40 | - | 96190..97137 [-], 948 | putative primase-like protein | |
| 41 | - | 97978..98337 [+], 360 | hypothetical protein | |
| 42 | - | 98347..98949 [+], 603 | hypothetical protein | |
| 43 | - | 99295..99831 [-], 537 | putative VC0181-like protein | |
| 44 | - | 99743..101512 [-], 1770 | putative VC180-like protein | |
| 45 | - | 101512..102810 [-], 1299 | putative VC0179-like protein | |
| 46 | - | 102810..103895 [-], 1086 | VC0178-like protein | |
| 47 | - | 104305..106023 [-], 1719 | hypothetical protein | |
| 48 | - | 106111..107004 [-], 894 | hypothetical protein | |
| 49 | - | 107020..107982 [-], 963 | putative patatin-like protein | |
| 50 | - | 108973..109299 [+], 327 | hypothetical protein | |
| 51 | - | 109923..111449 [+], 1527 | hypothetical protein | |
| 52 | - | 111555..112727 [-], 1173 | hypothetical protein | |
