
| 37 | |
| ICEPalBan1 | |
| ICEO_0000014 | |
| Providencia alcalifaciens Ban1 | |
| 96676 | |
| 47.09 | |
| prfC | |
| Antibiotic resistance genes: floR, strBA, sul2, dfrA1; Toxin-antitoxin system | |
| - | |
| GQ463139 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..96676 | |
| coordinates: 5905..6011; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 39381..41531; Locus tag: ICEPALBAN1_0039; Family: MOBH |
The graph information of ICEPalBan1 components from GQ463139 | |||||
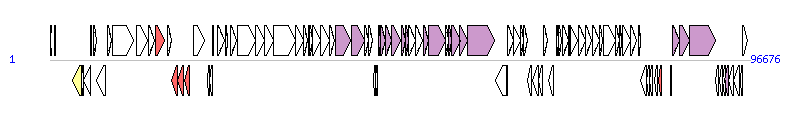 | |||||
| Complete gene list of ICEPalBan1 from GQ463139 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | ICEPALBAN1_0001 | 4..126 [+], 123 | peptide chain release factor 3 (RF-3) | ||
| 2 | ICEPALBAN1_0002 | 616..753 [+], 138 | conserved domain protein | ||
| 3 | ICEPALBAN1_0005 | 3046..4287 [-], 1242 | integrase | Integrase | |
| 4 | ICEPALBAN1_0006 | 4289..4558 [-], 270 | conserved hypothetical protein | ||
| 5 | ICEPALBAN1_0007 | 4561..5535 [-], 975 | conserved hypothetical protein | ||
| 6 | ICEPALBAN1_0008 | 5631..5798 [+], 168 | hypothetical protein | ||
| 7 | ICEPALBAN1_0009 | 6000..6443 [+], 444 | conserved hypothetical protein | ||
| 8 | ICEPALBAN1_0010 | 6454..7626 [-], 1173 | protein UmuC | ||
| 9 | ICEPALBAN1_0011 | 7910..8503 [+], 594 | transposase, IS3 family, interruption | ||
| 10 | ICEPALBAN1_0012 | 8617..11595 [+], 2979 | transposase | ||
| 11 | ICEPALBAN1_0013 | 12013..13506 [+], 1494 | iscr2 transposase | ||
| 12 | ICEPALBAN1_0014 | 13537..14421 [+], 885 | conserved hypothetical protein | ||
| 13 | ICEPALBAN1_0015 | 14638..15852 [+], 1215 | florfenicol/chloramphenicol resistance protein | AR | |
| 14 | ICEPALBAN1_0018 | 16296..16835 [+], 540 | truncated transposase | ||
| 15 | ICEPALBAN1_0017 | 16807..17637 [-], 831 | streptomycin resistance protein B | AR | |
| 16 | ICEPALBAN1_0019 | 17643..18446 [-], 804 | streptomycin 3''-kinase | AR | |
| 17 | folP | 18507..19322 [-], 816 | dihydropteroate synthase | AR | |
| 18 | ICEPALBAN1_0021 | 19793..21406 [+], 1614 | TnpA | ||
| 19 | ICEPALBAN1_0022 | 21749..22018 [-], 270 | protein UmuC | ||
| 20 | ICEPALBAN1_0023 | 22026..22475 [-], 450 | protein UmuD | ||
| 21 | ICEPALBAN1_0024 | 22474..22626 [+], 153 | hypothetical protein | ||
| 22 | ICEPALBAN1_0025 | 23131..24036 [+], 906 | exonuclease, RNase T and DNA polymerase III | ||
| 23 | ICEPALBAN1_0026 | 24250..24549 [+], 300 | conserved hypothetical protein | ||
| 24 | ICEPALBAN1_0027 | 24867..25790 [+], 924 | conserved hypothetical protein | ||
| 25 | ICEPALBAN1_0028 | 25904..28333 [+], 2430 | N-6 DNA methylase | ||
| 26 | ICEPALBAN1_0029 | 28333..29643 [+], 1311 | type I restriction-modification protein | ||
| 27 | ICEPALBAN1_0030 | 29653..30816 [+], 1164 | conserved hypothetical protein | ||
| 28 | ICEPALBAN1_0031 | 30831..33848 [+], 3018 | type I restriction-modification system restriction subunit | ||
| 29 | ICEPALBAN1_0032 | 33871..34956 [+], 1086 | conserved hypothetical protein | ||
| 30 | ICEPALBAN1_0033 | 34979..35530 [+], 552 | conserved hypothetical protein | ||
| 31 | ICEPALBAN1_0034 | 35660..35833 [+], 174 | conserved hypothetical protein | ||
| 32 | ICEPALBAN1_0035 | 35835..36203 [+], 369 | membrane-bound metal-dependent hydrolase | ||
| 33 | ICEPALBAN1_0036 | 36213..37367 [+], 1155 | conserved hypothetical protein | ||
| 34 | ICEPALBAN1_0037 | 37360..38550 [+], 1191 | conserved hypothetical protein | ||
| 35 | ICEPALBAN1_0038 | 38551..39264 [+], 714 | conserved hypothetical protein | ||
| 36 | ICEPALBAN1_0039 | 39381..41531 [+], 2151 | conjugative relaxase | Relaxase, MOBH Family | |
| 37 | ICEPALBAN1_0040 | 41579..43399 [+], 1821 | conjugative coupling factor | TraD_F, T4SS component | |
| 38 | ICEPALBAN1_0041 | 43409..43969 [+], 561 | conserved hypothetical protein | ||
| 39 | ICEPALBAN1_0042 | 43956..44591 [+], 636 | putative conjugation coupling factor | ||
| 40 | ICEPALBAN1_0043 | 44617..44895 [-], 279 | transcriptional regulator, XRE family | ||
| 41 | ICEPALBAN1_0044 | 44892..45215 [-], 324 | conserved hypothetical protein | ||
| 42 | traL | 45396..45677 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component | |
| 43 | ICEPALBAN1_0046 | 45674..46300 [+], 627 | sex pilus assembly | TraE_F, T4SS component | |
| 44 | ICEPALBAN1_0047 | 46284..47180 [+], 897 | TraK | TraK_F, T4SS component | |
| 45 | ICEPALBAN1_0048 | 47183..48472 [+], 1290 | sex pilus assembly | TraB_F, T4SS component | |
| 46 | ICEPALBAN1_0049 | 48547..49119 [+], 573 | sex pilus assembly | TraV_F, T4SS component | |
| 47 | ICEPALBAN1_0050 | 49116..49502 [+], 387 | TraA | TraA_F, T4SS component | |
| 48 | ICEPALBAN1_0051 | 49573..50514 [+], 942 | conserved hypothetical protein | ||
| 49 | ICEPALBAN1_0052 | 50507..51445 [+], 939 | ync | ||
| 50 | ICEPALBAN1_0053 | 51577..52269 [+], 693 | DsbC | TrbB_I, T4SS component | |
| 51 | traC | 52269..54668 [+], 2400 | type-IV secretion system protein TraC | TraC_F, T4SS component | |
| 52 | ICEPALBAN1_0055 | 54661..55008 [+], 348 | conserved hypothetical protein | ||
| 53 | ICEPALBAN1_0056 | 54992..55504 [+], 513 | conjugation signal peptidase | TraF, T4SS component | |
| 54 | ICEPALBAN1_0057 | 55515..56639 [+], 1125 | sex pilus assembly | TraW_F, T4SS component | |
| 55 | ICEPALBAN1_0058 | 56623..57651 [+], 1029 | sex pilus assembly | TraU_F, T4SS component | |
| 56 | ICEPALBAN1_0059 | 57654..61346 [+], 3693 | mating pair stabilization | TraN_F, T4SS component | |
| 57 | ICEPALBAN1_0060 | 61571..62995 [-], 1425 | transcriptional regulator containing a DNA-binding HTH domain and an aminotransferase domain | ||
| 58 | ICEPALBAN1_0061 | 63035..63175 [-], 141 | hypothetical protein | ||
| 59 | ICEPALBAN1_0062 | 63190..64068 [+], 879 | phenazine biosynthesis protein PhzF family | ||
| 60 | ICEPALBAN1_0063 | 64041..65036 [+], 996 | protein of unknown function DUF6, transmembrane | ||
| 61 | ICEPALBAN1_0064 | 65033..65428 [+], 396 | endoribonuclease L-PSP, putative | ||
| 62 | ICEPALBAN1_0065 | 65453..65914 [+], 462 | YbaK/prolyl-tRNA synthetase associated region | ||
| 63 | ICEPALBAN1_0066 | 65959..66552 [-], 594 | maltose O-acetyltransferase (maltose transacetylase) | ||
| 64 | panB | 66607..67452 [-], 846 | 3-methyl-2-oxobutanoate hydroxymethyltransferase | ||
| 65 | ICEPALBAN1_0068 | 67461..68084 [-], 624 | lysine exporter protein | ||
| 66 | ICEPALBAN1_0069 | 68213..68665 [+], 453 | transcriptional regulator | ||
| 67 | ICEPALBAN1_0070 | 68913..69515 [-], 603 | conserved hypothetical protein | ||
| 68 | ICEPALBAN1_0071 | 69883..70209 [+], 327 | conserved hypothetical protein | ||
| 69 | ICEPALBAN1_0072 | 70225..70662 [+], 438 | single-strand binding protein family | ||
| 70 | ICEPALBAN1_0073 | 70723..71541 [+], 819 | phage recombination protein Bet | ||
| 71 | ICEPALBAN1_0074 | 71624..71767 [+], 144 | hypothetical protein | ||
| 72 | ICEPALBAN1_0075 | 71828..72844 [+], 1017 | YqaJ viral recombinase family | ||
| 73 | ICEPALBAN1_0076 | 73054..74013 [+], 960 | ATPase associated with various cellular activities, AAA_5 | ||
| 74 | ICEPALBAN1_0077 | 74013..74780 [+], 768 | conserved hypothetical protein | ||
| 75 | ICEPALBAN1_0078 | 74879..75832 [+], 954 | conserved hypothetical protein | ||
| 76 | ICEPALBAN1_0079 | 75894..76334 [+], 441 | conserved hypothetical protein | ||
| 77 | ICEPALBAN1_0080 | 76404..78059 [+], 1656 | von Willebrand factor, type A | ||
| 78 | radC | 78144..78641 [+], 498 | DNA repair protein RadC | ||
| 79 | ICEPALBAN1_0082 | 78641..78982 [+], 342 | conserved hypothetical protein | ||
| 80 | ICEPALBAN1_0083 | 79074..80147 [+], 1074 | phage P4 alpha, zinc-binding domain protein | ||
| 81 | ICEPALBAN1_0084 | 80237..80944 [+], 708 | conserved hypothetical protein | ||
| 82 | ICEPALBAN1_0086 | 81216..81620 [+], 405 | glyoxalase/bleomycin resistance protein/dioxygenase | ||
| 83 | ICEPALBAN1_0085 | 81617..82318 [-], 702 | conserved hypothetical protein | ||
| 84 | ICEPALBAN1_0087 | 82418..82780 [-], 363 | conserved hypothetical protein | ||
| 85 | ICEPALBAN1_0088 | 82851..83261 [-], 411 | membrane associated protein containing xre-family DNA-binding HTH domain | ||
| 86 | ICEPALBAN1_0089 | 83459..83884 [-], 426 | conserved hypothetical protein | ||
| 87 | folA | 83989..84462 [-], 474 | dihydrofolate reductase | AR | |
| 88 | ICEPALBAN1_0091 | 85680..85820 [-], 141 | hypothetical protein | ||
| 89 | ICEPALBAN1_0093 | 85966..86910 [+], 945 | conserved hypothetical protein | TraF_F, T4SS component | |
| 90 | ICEPALBAN1_0094 | 86913..88301 [+], 1389 | sex pilus assembly | TraH_F, T4SS component | |
| 91 | ICEPALBAN1_0095 | 88305..91874 [+], 3570 | sex pilus assembly | TraG_F, T4SS component | |
| 92 | ICEPALBAN1_0096 | 91907..92338 [-], 432 | EexR2 | ||
| 93 | ICEPALBAN1_0097 | 92396..92929 [-], 534 | transcriptional activator | ||
| 94 | ICEPALBAN1_0098 | 92926..93225 [-], 300 | SetD | ||
| 95 | ICEPALBAN1_0099 | 93222..93770 [-], 549 | lytic transglycosylase, catalytic | Orf169_F, T4SS component | |
| 96 | ICEPALBAN1_0100 | 93757..94419 [-], 663 | conserved hypothetical protein | ||
| 97 | ICEPALBAN1_0101 | 94406..95275 [-], 870 | conserved hypothetical protein | ||
| 98 | ICEPALBAN1_0102 | 95331..95582 [-], 252 | SetQ | ||
| 99 | ICEPALBAN1_0103 | 95700..96347 [+], 648 | HTH-type transcriptional regulator for conjugative element R391 | ||
|
(1) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |
|
(2) Bordeleau E; Brouillette E; Robichaud N; Burrus V (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol. 12(2):510-23. [PubMed:19888998] |
|
(3) Marrero J; Waldor MK (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 189(8):3302-5. [PubMed:17307849] |