
| 23 | |
| ICEVchMex1 | |
| ICEO_0000011 | |
| Vibrio cholerae non O1-O139 Mex1 | |
| 83194 | |
| 46.73 | |
| prfC | |
| Resistant to ampicillin; restriction modification system | |
| - | |
| GQ463143 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..83194 | |
| coordinates: 5686..5792; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAA | |
| coordinates: 19499..21649; Locus tag: ICEVCHMEX1_0020; Family: MOBH |
The graph information of ICEVchMex1 components from GQ463143 | |||||
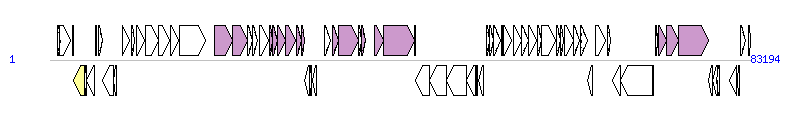 | |||||
| Complete gene list of ICEVchMex1 from GQ463143 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | ICEVCHMEX1_0001 | 872..1192 [+], 321 | transcriptional regulator, XRE family | ||
| 2 | ICEVCHMEX1_0002 | 1185..2399 [+], 1215 | HipA domain protein | ||
| 3 | ICEVCHMEX1_0003 | 2616..2810 [+], 195 | conserved domain protein | ||
| 4 | ICEVCHMEX1_0004 | 2827..4068 [-], 1242 | integrase | Integrase | |
| 5 | ICEVCHMEX1_0005 | 4070..4339 [-], 270 | conserved hypothetical protein | ||
| 6 | ICEVCHMEX1_0006 | 4342..5316 [-], 975 | conserved hypothetical protein | ||
| 7 | ICEVCHMEX1_0007 | 5412..5579 [+], 168 | hypothetical protein | ||
| 8 | ICEVCHMEX1_0008 | 5781..6224 [+], 444 | conserved hypothetical protein | ||
| 9 | ICEVCHMEX1_0009 | 6235..7554 [-], 1320 | protein UmuC | ||
| 10 | ICEVCHMEX1_0010 | 7511..7960 [-], 450 | protein UmuD | ||
| 11 | ICEVCHMEX1_0011 | 8600..9505 [+], 906 | exonuclease, RNase T and DNA polymerase III | ||
| 12 | ICEVCHMEX1_0012 | 9719..10018 [+], 300 | conserved hypothetical protein | ||
| 13 | ICEVCHMEX1_0013 | 10336..11259 [+], 924 | conserved hypothetical protein | ||
| 14 | hsdM | 11329..12903 [+], 1575 | type I restriction-modification system, M subunit | ||
| 15 | ICEVCHMEX1_0015 | 12900..14222 [+], 1323 | specificity subunit | ||
| 16 | ICEVCHMEX1_0016 | 14311..15312 [+], 1002 | filamentation induced by cAMP protein Fic | ||
| 17 | ICEVCHMEX1_0017 | 15376..18489 [+], 3114 | type I site-specific deoxyribonuclease, R subunit | ||
| 18 | ICEVCHMEX1_0020 | 19499..21649 [+], 2151 | conjugative relaxase | Relaxase, MOBH Family | |
| 19 | ICEVCHMEX1_0021 | 21698..23518 [+], 1821 | conjugative coupling factor | TraD_F, T4SS component | |
| 20 | ICEVCHMEX1_0022 | 23528..24088 [+], 561 | conserved hypothetical protein | ||
| 21 | ICEVCHMEX1_0023 | 24075..24710 [+], 636 | putative conjugation coupling factor | ||
| 22 | ICEVCHMEX1_0024 | 24848..25954 [+], 1107 | filamentation induced by cAMP protein Fic | ||
| 23 | traL | 26144..26425 [+], 282 | type IV conjugative transfer system protein TraL | TraL_F, T4SS component | |
| 24 | ICEVCHMEX1_0026 | 26422..27048 [+], 627 | sex pilus assembly | TraE_F, T4SS component | |
| 25 | ICEVCHMEX1_0027 | 27032..27931 [+], 900 | TraK | TraK_F, T4SS component | |
| 26 | ICEVCHMEX1_0028 | 27931..29220 [+], 1290 | sex pilus assembly | TraB_F, T4SS component | |
| 27 | ICEVCHMEX1_0029 | 29295..29867 [+], 573 | sex pilus assembly | TraV_F, T4SS component | |
| 28 | ICEVCHMEX1_0030 | 29864..30250 [+], 387 | TraA | TraA_F, T4SS component | |
| 29 | ICEVCHMEX1_0031 | 30291..30797 [-], 507 | acetyltransferase, gnat family | ||
| 30 | ICEVCHMEX1_0032 | 30788..31054 [-], 267 | conserved hypothetical protein | ||
| 31 | ICEVCHMEX1_0033 | 31209..31634 [-], 426 | conserved hypothetical protein | ||
| 32 | ICEVCHMEX1_0035 | 32567..33361 [+], 795 | aminoglycoside 3'-phosphotransferase | ||
| 33 | ICEVCHMEX1_0036 | 33571..34263 [+], 693 | DsbC | TrbB_I, T4SS component | |
| 34 | traC | 34264..36663 [+], 2400 | type-IV secretion system protein TraC | TraC_F, T4SS component | |
| 35 | ICEVCHMEX1_0038 | 36656..37003 [+], 348 | conserved hypothetical protein | ||
| 36 | ICEVCHMEX1_0039 | 36987..37499 [+], 513 | conjugation signal peptidase | TraF, T4SS component | |
| 37 | ICEVCHMEX1_0041 | 38618..39646 [+], 1029 | sex pilus assembly | TraU_F, T4SS component | |
| 38 | ICEVCHMEX1_0042 | 39649..43341 [+], 3693 | mating pair stabilization | TraN_F, T4SS component | |
| 39 | ICEVCHMEX1_0044 | 43354..43497 [+], 144 | hypothetical protein | ||
| 40 | ICEVCHMEX1_0043 | 43483..45159 [-], 1677 | UvrD/REP helicase | ||
| 41 | ICEVCHMEX1_0045 | 45156..47081 [-], 1926 | ATP-dependent endonuclease of the OLD family | ||
| 42 | ICEVCHMEX1_0046 | 47159..49537 [-], 2379 | peptidase, S8A | ||
| 43 | ICEVCHMEX1_0047 | 49553..50524 [-], 972 | ATPase, AAA family | ||
| 44 | ICEVCHMEX1_0048 | 50635..50814 [-], 180 | conserved hypothetical protein | ||
| 45 | ICEVCHMEX1_0049 | 50948..51550 [-], 603 | conserved hypothetical protein | ||
| 46 | ICEVCHMEX1_0050 | 51918..52244 [+], 327 | conserved hypothetical protein | ||
| 47 | ICEVCHMEX1_0051 | 52260..52679 [+], 420 | single-strand binding protein family | ||
| 48 | ICEVCHMEX1_0052 | 52759..53577 [+], 819 | phage recombination protein Bet | ||
| 49 | ICEVCHMEX1_0053 | 53659..53802 [+], 144 | hypothetical protein | ||
| 50 | ICEVCHMEX1_0054 | 53864..54880 [+], 1017 | YqaJ viral recombinase family | ||
| 51 | ICEVCHMEX1_0055 | 55090..56049 [+], 960 | ATPase associated with various cellular activities, AAA_5 | ||
| 52 | ICEVCHMEX1_0056 | 56049..56816 [+], 768 | conserved hypothetical protein | ||
| 53 | ICEVCHMEX1_0057 | 56915..57868 [+], 954 | conserved hypothetical protein | ||
| 54 | ICEVCHMEX1_0058 | 57930..58370 [+], 441 | conserved hypothetical protein | ||
| 55 | ICEVCHMEX1_0059 | 58440..60095 [+], 1656 | von Willebrand factor, type A | ||
| 56 | radC | 60178..60675 [+], 498 | DNA repair protein RadC | ||
| 57 | ICEVCHMEX1_0061 | 60675..61016 [+], 342 | conserved hypothetical protein | ||
| 58 | ICEVCHMEX1_0062 | 61108..62181 [+], 1074 | phage P4 alpha, zinc-binding domain protein | ||
| 59 | ICEVCHMEX1_0063 | 62271..62978 [+], 708 | conserved hypothetical protein | ||
| 60 | ICEVCHMEX1_0064 | 63001..63126 [+], 126 | hypothetical protein | ||
| 61 | ICEVCHMEX1_0065 | 63095..63847 [+], 753 | conserved hypothetical protein | ||
| 62 | ICEVCHMEX1_0066 | 63902..64438 [-], 537 | conserved hypothetical protein | ||
| 63 | ICEVCHMEX1_0067 | 64799..66070 [+], 1272 | pas/pac/ggdef-domain containing protein | ||
| 64 | ICEVCHMEX1_0068 | 66244..66669 [+], 426 | integrase, catalytic region | ||
| 65 | ICEVCHMEX1_0069 | 66830..67759 [-], 930 | response regulator receiver protein | ||
| 66 | ICEVCHMEX1_0070 | 67765..71571 [-], 3807 | multi-sensor hybrid histidine kinase | ||
| 67 | ICEVCHMEX1_0071 | 71572..71688 [-], 117 | hypothetical protein | ||
| 68 | ICEVCHMEX1_0072 | 72019..72165 [+], 147 | hypothetical protein | ||
| 69 | ICEVCHMEX1_0073 | 72311..73255 [+], 945 | conserved hypothetical protein | TraF_F, T4SS component | |
| 70 | ICEVCHMEX1_0074 | 73258..74646 [+], 1389 | sex pilus assembly | TraH_F, T4SS component | |
| 71 | ICEVCHMEX1_0075 | 74650..78219 [+], 3570 | sex pilus assembly | TraG_F, T4SS component | |
| 72 | ICEVCHMEX1_0076 | 78252..78683 [-], 432 | EexR3 | ||
| 73 | ICEVCHMEX1_0077 | 78741..79274 [-], 534 | transcriptional activator | ||
| 74 | ICEVCHMEX1_0078 | 79271..79570 [-], 300 | SetD | ||
| 75 | ICEVCHMEX1_0083 | 80759..81628 [-], 870 | conserved hypothetical protein | ||
| 76 | ICEVCHMEX1_0084 | 81684..81935 [-], 252 | SetQ | ||
| 77 | ICEVCHMEX1_0085 | 82053..82700 [+], 648 | HTH-type transcriptional regulator for conjugative element R391 | ||
| 78 | ICEVCHMEX1_0086 | 82957..83194 [+], 238 | peptide termination factor | ||
|
(1) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |
|
(2) Bordeleau E; Brouillette E; Robichaud N; Burrus V (2010). Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol. 12(2):510-23. [PubMed:19888998] |
|
(3) Marrero J; Waldor MK (2007). The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 189(8):3302-5. [PubMed:17307849] |
|
(4) Burrus V; Quezada-Calvillo R; Marrero J; Waldor MK (2006). SXT-related integrating conjugative element in New World Vibrio cholerae. Appl Environ Microbiol. 72(4):3054-7. [PubMed:16598018] |