
| 15 | |
| ICEVchBan8 | |
| Vibrio cholerae MZO-3 | |
| 103382 | |
| 41.8 | |
| tRNA-Ser | |
| Toxin-antitoxin system | |
| - | |
| JQ345361 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..103382 | |
| coordinates: 6385..6491; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAA | |
| - |
The graph information of ICEVchBan8 components from JQ345361 | |||||
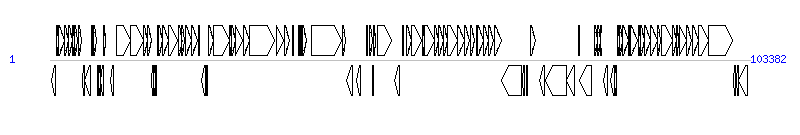 | |||||
| Complete gene list of ICEVchBan8 from JQ345361 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 178..825 [-], 648 | putative cI prophage repressor protein | ||
| 2 | - | 943..1194 [+], 252 | hypothetical protein | ||
| 3 | - | 1250..2119 [+], 870 | hypothetical protein | ||
| 4 | - | 2106..2768 [+], 663 | hypothetical protein | ||
| 5 | - | 2755..3303 [+], 549 | hypothetical protein | ||
| 6 | - | 3300..3599 [+], 300 | hypothetical protein | ||
| 7 | - | 3596..4129 [+], 534 | hypothetical protein | ||
| 8 | - | 4184..4615 [+], 432 | hypothetical protein | ||
| 9 | - | 4769..5038 [-], 270 | hypothetical protein | ||
| 10 | - | 5041..6015 [-], 975 | rod shape determination protein | ||
| 11 | - | 6111..6233 [+], 123 | hypothetical protein | ||
| 12 | - | 6480..6923 [+], 444 | hypothetical protein | ||
| 13 | - | 7061..7180 [-], 120 | error-prone, lesion bypass DNA polymerase V | ||
| 14 | - | 7180..7479 [-], 300 | error-prone, lesion bypass DNA polymerase V | ||
| 15 | - | 7448..7897 [-], 450 | error-prone repair protein | ||
| 16 | - | 7896..8153 [+], 258 | hypothetical protein | ||
| 17 | - | 8935..9408 [-], 474 | transposase | ||
| 18 | - | 9756..11906 [+], 2151 | conjugative transfer protein TraI | ||
| 19 | - | 11955..13775 [+], 1821 | IncF plasmid conjugative transfer protein TraD | ||
| 20 | - | 13785..14345 [+], 561 | conjugative transfer protein 234 | ||
| 21 | - | 14332..14967 [+], 636 | conjugative transfer protein s043 | ||
| 22 | - | 14993..15271 [-], 279 | hypothetical protein | ||
| 23 | - | 15268..15591 [-], 324 | hypothetical protein | ||
| 24 | - | 15605..15736 [-], 132 | hypothetical protein | ||
| 25 | - | 15841..16053 [+], 213 | IncF plasmid conjugative transfer pilus assembly protein TraL | ||
| 26 | - | 16050..16676 [+], 627 | IncF plasmid conjugative transfer pilus assembly protein TraE | ||
| 27 | - | 16660..17556 [+], 897 | IncF plasmid conjugative transfer pilus assembly protein TraK | ||
| 28 | - | 17559..18848 [+], 1290 | IncF plasmid conjugative transfer pilus assembly protein TraB | ||
| 29 | - | 18923..19495 [+], 573 | conjugative transfer protein TraV | ||
| 30 | - | 19492..19878 [+], 387 | conjugative transfer protein TraA | ||
| 31 | - | 20056..20889 [+], 834 | Ynd | ||
| 32 | - | 20882..21820 [+], 939 | Ync | ||
| 33 | - | 22004..22147 [+], 144 | hypothetical protein | ||
| 34 | - | 22343..22999 [-], 657 | hypothetical protein | ||
| 35 | - | 23043..23189 [-], 147 | hypothetical protein | ||
| 36 | - | 23431..24123 [+], 693 | thiol:disulfide | ||
| 37 | - | 24123..26522 [+], 2400 | IncF plasmid conjugative transfer pilus assembly protein TraC | ||
| 38 | - | 26515..26862 [+], 348 | conjugative transfer protein 345 | ||
| 39 | - | 26846..27358 [+], 513 | conjugative signal peptidase | ||
| 40 | - | 27369..28493 [+], 1125 | IncF plasmid conjugative transfer pilus assembly protein TraW | ||
| 41 | - | 28525..29505 [+], 981 | IncF plasmid conjugative transfer pilus assembly protein TraU | ||
| 42 | - | 29508..33206 [+], 3699 | IncF plasmid conjugative transfer protein TraN | ||
| 43 | - | 33384..34559 [+], 1176 | FOG: TPR repeat protein | ||
| 44 | - | 34573..35418 [+], 846 | outer membrane lipoprotein A precursor | ||
| 45 | - | 35745..35894 [+], 150 | hypothetical protein | ||
| 46 | - | 36729..36896 [+], 168 | ABC transporter, periplasmic substrate-binding protein | ||
| 47 | - | 36928..37185 [+], 258 | hypothetical protein | ||
| 48 | - | 37230..37883 [+], 654 | transposase and inactivated derivative | ||
| 49 | - | 38673..43061 [+], 4389 | Accessory colonization factor AcfD precursor | ||
| 50 | - | 43128..43682 [+], 555 | hypothetical protein | ||
| 51 | - | 43778..44698 [-], 921 | transposase | ||
| 52 | - | 45230..45859 [-], 630 | Accessory colonization factor AcfA | ||
| 53 | - | 46732..46902 [+], 171 | hypothetical protein | ||
| 54 | - | 47188..47571 [+], 384 | transthyretin family protein | ||
| 55 | - | 47621..47743 [-], 123 | hypothetical protein | ||
| 56 | - | 47716..48099 [+], 384 | protein of unknown function DUF336 | ||
| 57 | - | 48409..50502 [+], 2094 | Surface protein Lk90-like protein | ||
| 58 | - | 50844..51629 [-], 786 | Beta-ketoadipate enol-lactone hydrolase | ||
| 59 | - | 51992..52144 [+], 153 | hypothetical protein | ||
| 60 | - | 52601..53332 [+], 732 | toxin co-regulated pilin A | ||
| 61 | - | 53384..54922 [+], 1539 | toxin co-regulated pilus biosynthesis protein B | ||
| 62 | - | 54941..55312 [+], 372 | toxin co-regulated pilus biosynthesis protein Q | ||
| 63 | - | 55319..56779 [+], 1461 | toxin co-regulated pilus biosynthesis protein C, outer membrane protein | ||
| 64 | - | 56781..57320 [+], 540 | hypothetical protein | ||
| 65 | - | 57317..58150 [+], 834 | toxin co-regulated pilus biosynthesis protein D | ||
| 66 | - | 58138..58671 [+], 534 | CofG | ||
| 67 | - | 58673..60205 [+], 1533 | toxin co-regulated pilus biosynthesis protein T, putative ATP-binding translocase of TcpA | ||
| 68 | - | 60246..61217 [+], 972 | toxin co-regulated pilus biosynthesis protein E, anchors TcpT to membrane | ||
| 69 | - | 61218..61982 [+], 765 | TCP pilin signal peptidase, TcpA processing | ||
| 70 | - | 62057..62734 [+], 678 | toxin co-regulated pilus biosynthesis protein P, transcriptional activator of ToxT promoter | ||
| 71 | - | 62946..63110 [+], 165 | hypothetical protein | ||
| 72 | - | 63198..63995 [+], 798 | TCP pilus virulence regulatory protein ToxT, transcription activator | ||
| 73 | - | 64000..64719 [+], 720 | hypothetical protein | ||
| 74 | - | 64748..65497 [+], 750 | Accessory colonization factor AcfC | ||
| 75 | - | 65598..66617 [+], 1020 | hypothetical protein | ||
| 76 | - | 66748..69669 [-], 2922 | Lipoprotein, ToxR-activated gene, TagA | ||
| 77 | - | 69736..70140 [-], 405 | methyl-accepting chemotaxis sensory transducer | ||
| 78 | - | 70328..70555 [-], 228 | hypothetical protein | ||
| 79 | - | 70973..71776 [+], 804 | transcriptional regulator, AraC family | ||
| 80 | - | 72305..73066 [-], 762 | putative regulatory protein | ||
| 81 | - | 73239..76304 [-], 3066 | AcrB/AcrD/AcrF family protein | ||
| 82 | - | 76310..77395 [-], 1086 | hypothetical protein | ||
| 83 | - | 78009..78146 [+], 138 | hypothetical protein | ||
| 84 | - | 78225..80030 [-], 1806 | CtxA | ||
| 85 | - | 80450..80824 [+], 375 | transposase | ||
| 86 | - | 80931..81284 [+], 354 | transposase | ||
| 87 | - | 81235..81357 [+], 123 | transposase | ||
| 88 | - | 81767..82393 [-], 627 | Probable signal peptide protein | ||
| 89 | - | 82760..83365 [-], 606 | hypothetical protein | ||
| 90 | - | 83602..83718 [-], 117 | hypothetical protein | ||
| 91 | - | 83775..84059 [+], 285 | hypothetical protein | ||
| 92 | - | 84075..84494 [+], 420 | Single-stranded DNA-binding protein | ||
| 93 | - | 84574..85392 [+], 819 | Recombination protein BET | ||
| 94 | - | 85474..85617 [+], 144 | hypothetical protein | ||
| 95 | - | 85678..86694 [+], 1017 | hypothetical protein | ||
| 96 | - | 86904..87182 [+], 279 | Aerobic cobaltochelatase CobS subunit | ||
| 97 | - | 87249..87863 [+], 615 | Aerobic cobaltochelatase CobS subunit | ||
| 98 | - | 87863..88630 [+], 768 | hypothetical protein | ||
| 99 | - | 88729..89682 [+], 954 | Cobalamine biosynthesis protein | ||
| 100 | - | 89744..90184 [+], 441 | hypothetical protein | ||
| 101 | - | 90254..91903 [+], 1650 | Plasmid associated gene product APECO1_O1R37 | ||
| 102 | - | 91986..92483 [+], 498 | DNA repair protein RadC | ||
| 103 | - | 92483..92824 [+], 342 | hypothetical protein | ||
| 104 | - | 92915..93982 [+], 1068 | Phage P4 alpha, zinc-binding domain protein | ||
| 105 | - | 94072..94776 [+], 705 | hypothetical protein | ||
| 106 | - | 94881..95819 [+], 939 | IncF plasmid conjugative transfer pilus assembly protein TraF | ||
| 107 | - | 95816..97204 [+], 1389 | IncF plasmid conjugative transfer pilus assembly protein TraH | ||
| 108 | - | 97208..100777 [+], 3570 | IncF plasmid conjugative transfer protein TraG | ||
| 109 | - | 100950..101261 [-], 312 | hypothetical protein | ||
| 110 | - | 101460..101648 [-], 189 | transcriptional regulator | ||
| 111 | - | 101731..102963 [-], 1233 | Phage integrase | ||
|
(1) Taviani E; Spagnoletti M; Ceccarelli D; Haley BJ; Hasan NA; Chen A; Colombo MM; Huq A; Colwell RR (2012). Genomic analysis of ICEVchBan8: An atypical genetic element in Vibrio cholerae. FEBS Lett. 586(11):1617-21. [PubMed:22673571] |
|
(2) Wozniak RA; Fouts DE; Spagnoletti M; Colombo MM; Ceccarelli D; Garriss G; Dery C; Burrus V; Waldor MK (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5(12):e1000786. [PubMed:20041216] |