The graph information of ICEEcoED1a-1 components from CU928162 |
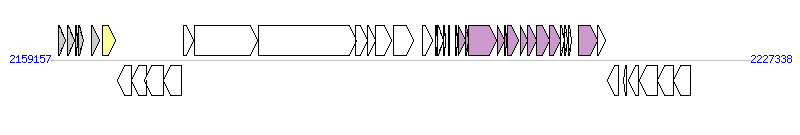 |
| Complete gene list of ICEEcoED1a-1 from CU928162 |
| # | Gene  | Coordinates [+/-], size (bp) | Product
(GenBank annotation) | *Reannotation  |
| 1 | yedZ | 2160015..2160650 [+], 636 | heme-molybdoenzyme heme-containing subunit YedZ; cytochrome b subunit | |
| 2 | yodA | 2160907..2161557 [+], 651 | conserved hypothetical protein; putative metal-binding protein | |
| 3 | ECED1_2239 | 2161667..2161816 [+], 150 | conserved hypothetical protein | |
| 4 | yodB | 2161900..2162430 [+], 531 | putative cytochrome | |
| 5 | yeeI | 2163183..2163980 [+], 798 | conserved hypothetical protein | |
| 6 | int | 2164318..2165580 [+], 1263 | integrase | Integrase |
| 7 | irp | 2165774..2167078 [-], 1305 | Salicylate synthase | |
| 8 | irp | 2167106..2168509 [-], 1404 | putative permease of yersiniabactin-iron transporter YbtX | |
| 9 | irp | 2168379..2170208 [-], 1830 | permease and ATP-binding protein of yersiniabactin-iron ABC transporter YbtQ | |
| 10 | irp | 2170168..2171970 [-], 1803 | permease and ATP-binding protein of yersiniabactin-iron ABC transporter YbtP | |
| 11 | ybtA | 2172137..2173096 [+], 960 | AraC type regulator of yersiniabactin gene cluster (HPI) | |
| 12 | irp | 2173269..2179394 [+], 6126 | High-molecular-weight nonribosomal peptide/polyketide synthetase 2 (HMWP2) | |
| 13 | irp | 2179482..2188973 [+], 9492 | High-molecular-weight nonribosomal peptide/polyketide synthetase 1 (HMWP1) | |
| 14 | irp | 2188910..2190070 [+], 1161 | Thiazolinyl-S-HMWP1 reductase YbtU | |
| 15 | irp | 2190067..2190870 [+], 804 | putative thioesterase YbtT | |
| 16 | irp | 2190874..2192451 [+], 1578 | salicyl-AMP ligase YbtE | |
| 17 | fyuA | 2192582..2194603 [+], 2022 | Yersiniabactin/pesticin outer membrane receptor (IRPC) | |
| 18 | ECED1_2255 | 2195417..2196400 [+], 984 | conserved hypothetical protein | |
| 19 | ECED1_2256 | 2196744..2196929 [+], 186 | conserved hypothetical protein | |
| 20 | ECED1_2257 | 2196914..2197495 [+], 582 | conserved hypothetical protein | |
| 21 | ECED1_2258 | 2197485..2197694 [+], 210 | putative lambdoid prophage protein from the DksA/TraR family with a zinc finger | |
| 22 | ECED1_2259 | 2197937..2198065 [+], 129 | conserved hypothetical protein | |
| 23 | ECED1_2260 | 2198634..2198867 [+], 234 | conserved hypothetical protein | |
| 24 | ECED1_2261 | 2198922..2199650 [+], 729 | putative type IV secretory pathway VirB1 component | VirB1, T4SS component |
| 25 | ECED1_2262 | 2199650..2199943 [+], 294 | putative type IV secretory pathway VirB2 component | VirB2, T4SS component |
| 26 | ECED1_2263 | 2199956..2202694 [+], 2739 | putative type IV secretory pathway VirB4 component | VirB4, T4SS component |
| 27 | ECED1_2264 | 2202712..2203419 [+], 708 | putative type IV secretory pathway VirB5 component | VirB5, T4SS component |
| 28 | ECED1_2265 | 2203427..2203669 [+], 243 | conserved hypothetical protein; putative exported protein | MagB05, T4SS component |
| 29 | ECED1_2266 | 2203673..2204746 [+], 1074 | putative type IV secretory pathway VirB6 component | VirB6, T4SS component |
| 30 | ECED1_2267 | 2204968..2205651 [+], 684 | putative type IV secretory pathway VirB8 component | VirB8, T4SS component |
| 31 | ECED1_2268 | 2205648..2206556 [+], 909 | putative type IV secretory pathway VirB9 component | VirB9, T4SS component |
| 32 | ECED1_2269 | 2206586..2207842 [+], 1257 | putative type IV secretory pathway VirB10 component | VirB10, T4SS component |
| 33 | ECED1_2270 | 2207832..2208857 [+], 1026 | putative type IV secretory pathway VirB11 component | VirB11, T4SS component |
| 34 | ECED1_2271 | 2208854..2209252 [+], 399 | conserved hypothetical protein; putative exported protein | |
| 35 | kikA | 2209288..2209593 [+], 306 | putative plasmid conjugation protein | |
| 36 | ECED1_2273 | 2209627..2209932 [+], 306 | conserved hypothetical protein | |
| 37 | ECED1_2274 | 2210612..2212501 [+], 1890 | putative MobB mobilization protein | VirD4, T4SS component |
| 38 | ECED1_2275 | 2212511..2213257 [+], 747 | putative MobC mobilization protein | |
| 39 | ECED1_2276 | 2213451..2214497 [-], 1047 | putative Antirestriction protein ardC | |
| 40 | ECED1_2277 | 2215060..2215323 [-], 264 | Transposase, ORF A, IS3 family, IS407 group | |
| 41 | ECED1_2278 | 2215465..2216523 [-], 1059 | conserved hypothetical protein | |
| 42 | ECED1_2279 | 2216529..2218313 [-], 1785 | conserved hypothetical protein | |
| 43 | ECED1_2280 | 2218310..2219875 [-], 1566 | putative Restriction modification system, type I similar to hsdS (fragment) | |
| 44 | ECED1_2281 | 2219872..2221545 [-], 1674 | putative HsdM; type I restriction modification enzyme methylase subunit | |
