
| 1069 | |
| ICEVchNig1 | |
| ICEO_0000254 | |
| Vibrio cholerae O1 VC833 | |
| 97043 | |
| 46.79 | |
| - | |
| Antibiotic resistance genes: floR, strAB, sul2, dfrA1 | |
| - | |
| KC886258 (complete ICE sequence in this GenBank file) | |
| - | |
| 528..97570 | |
| coordinates: 5869..5975; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| - |
The graph information of ICEVchNig1 components from KC886258 | |||||
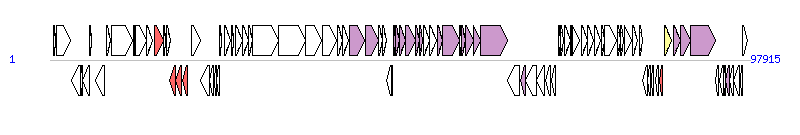 | |||||
| Complete gene list of ICEVchNig1 from KC886258 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | K531_00005 | 528..716 [+], 189 | DNA-binding protein | ||
| 2 | K531_00010 | 943..2880 [+], 1938 | hypothetical protein | ||
| 3 | K531_00015 | 3010..4251 [-], 1242 | phage Integrase | ||
| 4 | K531_00020 | 4253..4522 [-], 270 | hypothetical protein | ||
| 5 | K531_00025 | 4525..5499 [-], 975 | rod shape determination protein | ||
| 6 | K531_00030 | 5595..5762 [+], 168 | hypothetical protein | ||
| 7 | K531_00035 | 6418..7590 [-], 1173 | DNA polymerase V | ||
| 8 | K531_00040 | 7874..8467 [+], 594 | transposase Tn3 | ||
| 9 | K531_00045 | 8581..11559 [+], 2979 | transposase Tn3 family protein | ||
| 10 | K531_00050 | 11674..11865 [+], 192 | RepA protein | ||
| 11 | K531_00055 | 11977..13470 [+], 1494 | InsA | ||
| 12 | K531_00060 | 13501..14385 [+], 885 | Type IV secretory pathway VirD2 component | ||
| 13 | K531_00065 | 14602..15816 [+], 1215 | FloR | AR | |
| 14 | K531_00070 | 15844..16149 [+], 306 | putative transcriptional regulator protein | ||
| 15 | K531_00075 | 16261..16800 [+], 540 | TnpA | ||
| 16 | K531_00080 | 16772..17608 [-], 837 | streptomycin 3''-kinase | AR | |
| 17 | K531_00085 | 17608..18411 [-], 804 | StrA | AR | |
| 18 | K531_00090 | 18472..19287 [-], 816 | dihydropteroate synthase | AR | |
| 19 | K531_00095 | 19758..21110 [+], 1353 | transposase Tn3 family protein | ||
| 20 | K531_00100 | 21102..22313 [-], 1212 | transposase mutator family protein | ||
| 21 | K531_00105 | 22371..22874 [-], 504 | MutL protein | ||
| 22 | K531_00110 | 23027..23296 [-], 270 | error-prone repair protein UmuC | ||
| 23 | K531_00115 | 23304..23753 [-], 450 | DNA polymerase V | ||
| 24 | K531_00120 | 23752..24009 [+], 258 | hypothetical protein | ||
| 25 | K531_00125 | 24393..25298 [+], 906 | DNA polymerase III subunit epsilon | ||
| 26 | K531_00130 | 25386..25685 [+], 300 | plasmid-related protein | ||
| 27 | K531_00135 | 26003..26923 [+], 921 | hypothetical protein | ||
| 28 | K531_00140 | 26941..27741 [+], 801 | hypothetical protein | ||
| 29 | K531_00145 | 27725..28273 [+], 549 | hypothetical protein | ||
| 30 | K531_00150 | 28286..31975 [+], 3690 | hypothetical protein | ||
| 31 | K531_00155 | 31980..35720 [+], 3741 | hypothetical protein | ||
| 32 | K531_00160 | 35733..38036 [+], 2304 | PglZ domain protein | ||
| 33 | K531_00165 | 38075..40126 [+], 2052 | ATP-dependent Lon protease | ||
| 34 | K531_00170 | 40153..40923 [+], 771 | hypothetical protein | ||
| 35 | K531_00175 | 40958..41773 [+], 816 | hypothetical protein | ||
| 36 | K531_00180 | 41895..44045 [+], 2151 | conjugal transfer pilus assembly protein TraI | ||
| 37 | K531_00185 | 44094..45914 [+], 1821 | conjugal transfer pilus assembly protein TraD | TraD_F, T4SS component | |
| 38 | K531_00190 | 45924..46484 [+], 561 | plasmid-related protein | ||
| 39 | K531_00195 | 46471..47106 [+], 636 | plasmid-related protein | ||
| 40 | K531_00200 | 47133..47720 [-], 588 | hypothetical protein | ||
| 41 | K531_00205 | 47734..47973 [-], 240 | plasmid-related protein | ||
| 42 | K531_00210 | 48009..48290 [+], 282 | IncF plasmid conjugative transfer pilus assembly protein TraL | TraL_F, T4SS component | |
| 43 | K531_00215 | 48287..48913 [+], 627 | IncF plasmid conjugative transfer pilus assembly protein TraE | TraE_F, T4SS component | |
| 44 | K531_00220 | 48897..49793 [+], 897 | conjugal transfer pilus assembly protein TraK | TraK_F, T4SS component | |
| 45 | K531_00225 | 49796..51085 [+], 1290 | conjugal transfer pilus assembly protein TraB | TraB_F, T4SS component | |
| 46 | K531_00230 | 51160..51732 [+], 573 | Conjugative transfer protein TraV | TraV_F, T4SS component | |
| 47 | K531_00235 | 51729..52115 [+], 387 | conjugal transfer pilus assembly protein TraA | TraA_F, T4SS component | |
| 48 | K531_00240 | 52294..53127 [+], 834 | Ynd | ||
| 49 | K531_00245 | 53120..54058 [+], 939 | plasmid-related protein | ||
| 50 | K531_00250 | 54190..54882 [+], 693 | thiol:disulfide interchange protein DsbC | TrbB_I, T4SS component | |
| 51 | K531_00255 | 54882..57281 [+], 2400 | conjugal transfer ATP-binding protein TraC | TraC_F, T4SS component | |
| 52 | K531_00260 | 57274..57621 [+], 348 | hypothetical protein | ||
| 53 | K531_00265 | 57605..58117 [+], 513 | conjugal transfer pilin signal peptidase TrbI | TraF, T4SS component | |
| 54 | K531_00270 | 58128..59252 [+], 1125 | conjugal transfer pilus assembly protein TraW | TraW_F, T4SS component | |
| 55 | K531_00275 | 59236..60264 [+], 1029 | conjugal transfer pilus assembly protein TraU | TraU_F, T4SS component | |
| 56 | K531_00280 | 60267..63959 [+], 3693 | conjugal transfer mating pair stabilization protein TraN | TraN_F, T4SS component | |
| 57 | K531_00285 | 64050..65666 [-], 1617 | SMC domain protein | ||
| 58 | K531_00290 | 65746..66501 [-], 756 | IstB ATP binding domain-containing protein | TrbB, T4SS component | |
| 59 | K531_00295 | 66491..67999 [-], 1509 | integrase catalytic subunit | ||
| 60 | K531_00300 | 68098..69141 [-], 1044 | hypothetical protein | ||
| 61 | K531_00305 | 69345..70028 [-], 684 | deoxyribonuclease I | ||
| 62 | K531_00310 | 70137..70739 [-], 603 | plasmid-related protein | ||
| 63 | K531_00315 | 71107..71433 [+], 327 | hypothetical protein | ||
| 64 | K531_00320 | 71449..71868 [+], 420 | single-strand DNA-binding protein | ||
| 65 | K531_00325 | 71948..72766 [+], 819 | phage recombination protein | ||
| 66 | K531_00330 | 72848..72991 [+], 144 | hypothetical protein | ||
| 67 | K531_00335 | 73052..74068 [+], 1017 | endonuclease-like phage-related protein | ||
| 68 | K531_00340 | 74278..75237 [+], 960 | cobaltochelatase CobS | ||
| 69 | K531_00345 | 75237..76004 [+], 768 | hypothetical protein | ||
| 70 | K531_00350 | 76103..77056 [+], 954 | cobalamine biosynthesis protein | ||
| 71 | K531_00355 | 77118..77558 [+], 441 | hypothetical protein | ||
| 72 | K531_00360 | 77628..79283 [+], 1656 | hypothetical protein | ||
| 73 | K531_00365 | 79368..79865 [+], 498 | DNA repair protein RadC | ||
| 74 | K531_00370 | 79865..80206 [+], 342 | hypothetical protein | ||
| 75 | K531_00375 | 80297..81370 [+], 1074 | hypothetical protein | ||
| 76 | K531_00380 | 81460..82167 [+], 708 | hypothetical protein | ||
| 77 | K531_00385 | 82439..82843 [+], 405 | glyoxalase/bleomycin resistance protein/dioxygenase | ||
| 78 | K531_00390 | 82840..83541 [-], 702 | hypothetical protein | ||
| 79 | K531_00395 | 83641..84003 [-], 363 | hypothetical protein | ||
| 80 | K531_00400 | 84074..84484 [-], 411 | DNA-binding HTH domain-protein | ||
| 81 | K531_00405 | 84682..85107 [-], 426 | hypothetical protein | ||
| 82 | K531_00410 | 85212..85685 [-], 474 | dihydrofolate reductase | AR | |
| 83 | K531_00415 | 85973..86932 [+], 960 | integron integrase | Integrase | |
| 84 | K531_00420 | 87189..88133 [+], 945 | conjugal transfer pilus assembly protein TraF | TraF_F, T4SS component | |
| 85 | K531_00425 | 88136..89524 [+], 1389 | conjugative transfer pilus assembly protein TraH | TraH_F, T4SS component | |
| 86 | K531_00430 | 89528..93097 [+], 3570 | conjugal transfer mating pair stabilization protein TraG | TraG_F, T4SS component | |
| 87 | K531_00435 | 93130..93561 [-], 432 | hypothetical protein | ||
| 88 | K531_00440 | 93619..94152 [-], 534 | flagellar transcriptional activator FlhC | ||
| 89 | K531_00445 | 94149..94448 [-], 300 | flagellar transcriptional activator FlhD | ||
| 90 | K531_00450 | 94445..94993 [-], 549 | lytic transglycosylase, catalytic | Orf169_F, T4SS component | |
| 91 | K531_00455 | 94980..95642 [-], 663 | hypothetical protein | ||
| 92 | K531_00460 | 95629..96498 [-], 870 | hypothetical protein | ||
| 93 | K531_00465 | 96554..96805 [-], 252 | hypothetical protein | ||
| 94 | K531_00470 | 96923..97570 [+], 648 | HTH-type transcriptional regulator for conjugative element SXT | ||
| (1) Marin MA; Fonseca EL; Andrade BN; Cabral AC; Vicente AC (2014). Worldwide occurrence of integrative conjugative element encoding multidrug resistance determinants in epidemic Vibrio cholerae O1. PLoS One. 9(9):e108728. [PubMed:25265418] |