
| 1064 | |
| ICEVchChnAHV1003 | |
| Vibrio cholerae AHV1003 | |
| 101856 | |
| 47.45 | |
| - | |
| Antibiotic resistance genes: dfrA1, bcr, sul2, strAB, tetAR, mphRK, mrx | |
| - | |
| KT151663 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..101856 | |
| coordinates: 95960..96066; oriTDB id: 200056 TTCCCCATCCAATTAACCCCTAAAGCCAAAACCACTATCCGTTTGGCTTTTGGATCGAAACGCCAAACGT AAAACTGCCGTCACAATCACTGTTTGGCGTCTCGATA | |
| coordinates: 53291..55441; Gene: traI; Family: MOBH |
The graph information of ICEVchChnAHV1003 components from KT151663 | |||||
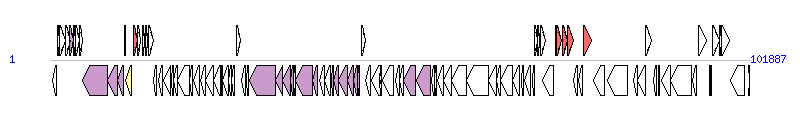 | |||||
| Complete gene list of ICEVchChnAHV1003 from KT151663 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | setR | 325..972 [-], 648 | SetR | ||
| 2 | - | 1090..1341 [+], 252 | hypothetical protein | ||
| 3 | - | 1397..2266 [+], 870 | hypothetical protein | ||
| 4 | - | 2253..2915 [+], 663 | hypothetical protein | ||
| 5 | - | 2902..3450 [+], 549 | hypothetical protein | Orf169_F, T4SS component | |
| 6 | setD | 3447..3746 [+], 300 | SetD | ||
| 7 | setC | 3743..4276 [+], 534 | SetC | ||
| 8 | eexR1 | 4334..4765 [+], 432 | EexR1 | ||
| 9 | traG | 4798..8367 [-], 3570 | plasmid conjugative transfer protein | TraG_F, T4SS component | |
| 10 | traH | 8371..9759 [-], 1389 | plasmid conjugative transfer pilus assembly protein trah | TraH_F, T4SS component | |
| 11 | traF | 9762..10706 [-], 945 | plasmid conjugative transfer pilus assembly protein traf | TraF_F, T4SS component | |
| 12 | - | 10852..10992 [+], 141 | hypothetical protein | ||
| 13 | intIPac | 10963..11922 [-], 960 | integrase | Integrase | |
| 14 | dhfR | 12210..12683 [+], 474 | dihydrofolate reductase | AR | |
| 15 | - | 12788..13213 [+], 426 | hypothetical protein | ||
| 16 | - | 13411..13821 [+], 411 | membrane associated protein | ||
| 17 | - | 13892..14254 [+], 363 | hypothetical protein | ||
| 18 | - | 14354..15055 [+], 702 | hypothetical protein | ||
| 19 | - | 15052..15456 [-], 405 | glyoxalase/bleomycin resistance protein/dioxygenase | ||
| 20 | - | 15728..16435 [-], 708 | hypothetical protein | ||
| 21 | - | 16525..17598 [-], 1074 | ATPase | ||
| 22 | - | 17689..18030 [-], 342 | hypothetical protein | ||
| 23 | radC | 18030..18527 [-], 498 | DNA repair protein | ||
| 24 | cobT | 18611..20266 [-], 1656 | cobalamin biosynthesis protein | ||
| 25 | - | 20336..20776 [-], 441 | hypothetical protein | ||
| 26 | - | 20838..21791 [-], 954 | hypothetical protein | ||
| 27 | - | 21890..22657 [-], 768 | hypothetical protein | ||
| 28 | cobS | 22657..23733 [-], 1077 | aerobic cobaltochelatase cobs subunit | ||
| 29 | - | 23826..24842 [-], 1017 | hypothetical protein | ||
| 30 | - | 24903..25046 [-], 144 | hypothetical protein | ||
| 31 | bet | 25128..25946 [-], 819 | recombination protein BET | ||
| 32 | ssd | 26026..26445 [-], 420 | single-stranded DNA-binding protein | ||
| 33 | - | 26461..26787 [-], 327 | hypothetical protein | ||
| 34 | - | 27155..27757 [+], 603 | hypothetical protein | ||
| 35 | - | 27848..28510 [-], 663 | hypothetical protein | ||
| 36 | - | 28541..28873 [-], 333 | hypothetical protein | ||
| 37 | traN | 29105..32797 [-], 3693 | plasmid conjugative transfer protein | TraN_F, T4SS component | |
| 38 | traU | 32800..33828 [-], 1029 | plasmid conjugative transfer pilus assembly protein trau | TraU_F, T4SS component | |
| 39 | traW | 33812..34936 [-], 1125 | plasmid conjugative transfer pilus assembly protein traw | TraW_F, T4SS component | |
| 40 | trhF | 34947..35459 [-], 513 | conjugative signal peptidase | TraF, T4SS component | |
| 41 | - | 35443..35790 [-], 348 | hypothetical protein | ||
| 42 | traC | 35783..38182 [-], 2400 | plasmid conjugative transfer pilus assembly protein trac | TraC_F, T4SS component | |
| 43 | - | 38182..38874 [-], 693 | thiol:disulfide protein | TrbB_I, T4SS component | |
| 44 | ync | 39253..39942 [-], 690 | ync | ||
| 45 | ynd | 39935..40768 [-], 834 | ynd | ||
| 46 | traA | 40946..41332 [-], 387 | conjugative transfer protein | TraA_F, T4SS component | |
| 47 | traV | 41329..41979 [-], 651 | conjugative transfer protein | TraV_F, T4SS component | |
| 48 | traB | 41976..43265 [-], 1290 | plasmid conjugative transfer pilus assembly protein trab | TraB_F, T4SS component | |
| 49 | traK | 43268..44164 [-], 897 | plasmid conjugative transfer pilus assembly protein trak | TraK_F, T4SS component | |
| 50 | traE | 44148..44774 [-], 627 | plasmid conjugative transfer pilus assembly protein trae | TraE_F, T4SS component | |
| 51 | traL | 44771..45052 [-], 282 | plasmid conjugative transfer pilus assembly protein tral | TraL_F, T4SS component | |
| 52 | - | 45341..45913 [+], 573 | hypothetical protein | ||
| 53 | iSPsy4 | 45992..46687 [-], 696 | ISPsy4 transposition helper protein | ||
| 54 | - | 46763..48034 [-], 1272 | hypothetical protein | ||
| 55 | - | 47994..48242 [-], 249 | hypothetical protein | ||
| 56 | tnp | 48402..49958 [-], 1557 | transposase | ||
| 57 | - | 50230..50865 [-], 636 | conjugative transfer protein | ||
| 58 | - | 50852..51412 [-], 561 | conjugative transfer protein | ||
| 59 | traD | 51422..53242 [-], 1821 | plasmid conjugative transfer protein | TraD_F, T4SS component | |
| 60 | traI | 53291..55441 [-], 2151 | conjugative transfer protein relaxase | Relaxase, MOBH Family | |
| 61 | - | 55561..56034 [-], 474 | hypothetical protein | ||
| 62 | - | 56034..56315 [-], 282 | hypothetical protein | ||
| 63 | mrr | 56409..57263 [-], 855 | mrr restriction system protein | ||
| 64 | mcrC | 57263..58390 [-], 1128 | putative McrC protein | ||
| 65 | - | 58390..60588 [-], 2199 | hypothetical protein | ||
| 66 | - | 60604..63768 [-], 3165 | type I restriction-modification system restriction subunit r | ||
| 67 | bstXI | 63773..64927 [-], 1155 | bstXI restriction endonuclease | ||
| 68 | - | 64924..65628 [-], 705 | type I restriction-modification system specificity subunit s | ||
| 69 | - | 65621..67336 [-], 1716 | type I restriction-modification system DNA-methyltransferase subunit m | ||
| 70 | - | 67345..68295 [-], 951 | hypothetical protein | ||
| 71 | - | 68617..68916 [-], 300 | hypothetical protein | ||
| 72 | - | 69004..69909 [-], 906 | DNA polymerase III epsilon subunit-like protein | ||
| 73 | - | 70292..70549 [-], 258 | hypothetical protein | ||
| 74 | umuD | 70548..70997 [+], 450 | error-prone repair protein | ||
| 75 | rumB | 71005..71274 [+], 270 | RumB | ||
| 76 | - | 71427..72149 [+], 723 | hypothetical protein | ||
| 77 | tnpA | 71617..73230 [-], 1614 | transposase | ||
| 78 | - | 73510..73692 [+], 183 | hypothetical protein | ||
| 79 | sul2 | 73701..74516 [+], 816 | dihydropteroate synthase | AR | |
| 80 | strA | 74577..75380 [+], 804 | aminoglycoside phosphotransferase | AR | |
| 81 | strB | 75386..76216 [+], 831 | streptomycin phosphotransferase | AR | |
| 82 | insA | 76188..76727 [-], 540 | transposase | ||
| 83 | tetR | 76906..77508 [-], 603 | TetR family transcriptional regulator | ||
| 84 | tetA | 77589..78791 [+], 1203 | tetracycline efflux protein | AR | |
| 85 | tnpB | 79170..80663 [-], 1494 | transposase | ||
| 86 | - | 81081..84059 [-], 2979 | transposase | ||
| 87 | mphR | 84876..85460 [-], 585 | MphR | ||
| 88 | mrx | 85460..86698 [-], 1239 | mrx protein | ||
| 89 | mphK | 86605..87621 [+], 1017 | macrolide phosphotransferase K | ||
| 90 | - | 87838..88299 [-], 462 | hypothetical protein | ||
| 91 | - | 88566..88694 [-], 129 | hypothetical protein | ||
| 92 | tnpA | 88962..90257 [-], 1296 | TnpA | ||
| 93 | tnp | 90376..93354 [-], 2979 | transposase | ||
| 94 | tnp | 93468..94061 [-], 594 | transposase | ||
| 95 | umuC | 94345..95517 [+], 1173 | error-prone lesion bypass DNA polymerase V | ||
| 96 | - | 95994..96128 [-], 135 | hypothetical protein | ||
| 97 | - | 96173..96340 [-], 168 | hypothetical protein | ||
| 98 | - | 96436..97410 [+], 975 | hypothetical protein | ||
| 99 | - | 97413..97682 [+], 270 | hypothetical protein | ||
| 100 | int | 97684..98925 [+], 1242 | integrase-like protein | ||
| 101 | mutL | 99055..101073 [-], 2019 | DNA mismatch repair protein | ||
| 102 | - | 101743..101871 [-], 129 | hypothetical protein | ||
| (1) Ryan MP; Armshaw P; O'Halloran JA; Pembroke JT (2017). Analysis and comparative genomics of R997, the first SXT/R391 integrative and conjugative element (ICE) of the Indian Sub-Continent. Sci Rep. 7(1):8562. [PubMed:28819148] |
|
(2) Wang R; Yu D; Yue J; Kan B (2016). Variations in SXT elements in epidemic Vibrio cholerae O1 El Tor strains in China. Sci Rep. 6:22733. [PubMed:26956038] |