
| 1044 | |
| ICEValHN396 | |
| Vibrio alginolyticus HN396 | |
| 86687 | |
| 46.49 | |
| tRNA-ser | |
| - | |
| - | |
| KT072770 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..86687 | |
| coordinates: 3663..3769; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAA | |
| coordinates: 27634..29784; Locus tag: ICEValHN396_019; Family: MOBH |
The graph information of ICEValHN396 components from KT072770 | |||||
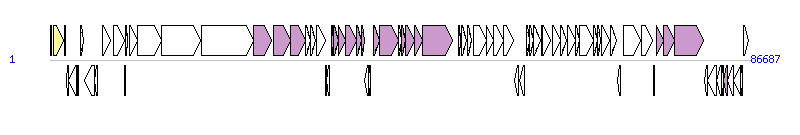 | |||||
| Complete gene list of ICEValHN396 from KT072770 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | ICEValHN396_001 | 15..134 [+], 120 | Hypothetical protein | ||
| 2 | int | 421..1653 [+], 1233 | Integrase | Integrase | |
| 3 | xis | 1738..1929 [+], 192 | Recombination directionality factor, Xis | ||
| 4 | s002 | 2045..2317 [-], 273 | Hypothetical protein, S002 | ||
| 5 | s003 | 2320..3294 [-], 975 | Rod shape determination protein,S003 | ||
| 6 | ICEValHN396_006 | 3457..3582 [-], 126 | Hypothetical protein | ||
| 7 | mobI | 3758..4201 [+], 444 | Hypothetical protein, MobI | ||
| 8 | rumB | 4269..5480 [-], 1212 | Error-prone repair protein, RumB | ||
| 9 | rumA | 5488..5937 [-], 450 | Error-prone repair protein, RumA | ||
| 10 | s024 | 6563..7468 [+], 906 | DNA polymerase III, S024 | ||
| 11 | ICEValHN396_011 | 7841..9220 [+], 1380 | Transposase | ||
| 12 | ICEValHN396_012 | 9217..9366 [-], 150 | Hypothetical protein | ||
| 13 | s025 | 9352..9540 [+], 189 | Hypothetical protein,S025 | ||
| 14 | s026 | 9859..10782 [+], 924 | Hypothetical protein, S026 | ||
| 15 | ICEValHN396_015 | 10884..13772 [+], 2889 | ATP-dependent helicase | ||
| 16 | ICEValHN396_016 | 13775..18646 [+], 4872 | Type II restriction enzyme, methylase subunits | ||
| 17 | ICEValHN396_017 | 18823..25245 [+], 6423 | DEAD/DEAH box helicase | ||
| 18 | ICEValHN396_018 | 25251..27386 [+], 2136 | Putative DNA helicase | TraI_F, T4SS component | |
| 19 | rraI | 27634..29784 [+], 2151 | Conjugative transfer protein, TraI | Relaxase, MOBH Family | |
| 20 | traD | 29832..31667 [+], 1836 | Conjugative transfer protein, TraD | TraD_F, T4SS component | |
| 21 | ICEValHN396_021 | 31677..32237 [+], 561 | Conjugative transfer protein, S091 | ||
| 22 | traJ | 32224..32859 [+], 636 | Conjugative transfer protein, TraJ | ||
| 23 | ICEValHN396_023 | 32997..34103 [+], 1107 | Fic family protein | ||
| 24 | ICEValHN396_024 | 34159..34293 [-], 135 | Hypothetical protein | ||
| 25 | hipA | 34299..34595 [-], 297 | HigA protein | ||
| 26 | traL | 34829..35110 [+], 282 | Conjugative transfer pilus assembly protein, TraL | TraL_F, T4SS component | |
| 27 | traE | 35119..35733 [+], 615 | Conjugative transfer pilus assembly protein, TraE | TraE_F, T4SS component | |
| 28 | traK | 35717..36613 [+], 897 | Conjugative transfer pilus assembly protein, TraK | TraK_F, T4SS component | |
| 29 | traB | 36616..37905 [+], 1290 | Conjugative transfer pilus assembly protein, TraB | TraB_F, T4SS component | |
| 30 | traV | 37902..38552 [+], 651 | Conjugative transfer protein, TraV | TraV_F, T4SS component | |
| 31 | traA | 38549..38935 [+], 387 | Conjugative transfer protein, TraA | TraA_F, T4SS component | |
| 32 | ICEValHN396_032 | 38976..39482 [-], 507 | Acetyltransferase | ||
| 33 | ICEValHN396_033 | 39473..39739 [-], 267 | Hypothetical protein | ||
| 34 | dsbC | 40109..40801 [+], 693 | Thiol:disulfide involved in conjugative transfer, dsbC | TrbB_I, T4SS component | |
| 35 | traC | 40802..43204 [+], 2403 | Conjugative transfer pilus assembly protein, TraC | TraC_F, T4SS component | |
| 36 | ICEValHN396_036 | 43197..43544 [+], 348 | Conjugative transfer protein 345 | ||
| 37 | trhF | 43528..44040 [+], 513 | Conjugative signal peptidase, TrhF | TraF, T4SS component | |
| 38 | traW | 44051..45175 [+], 1125 | Conjugative transfer pilus assembly protein, TraW | TraW_F, T4SS component | |
| 39 | traU | 45159..46187 [+], 1029 | Conjugative transfer protein, TraU | TraU_F, T4SS component | |
| 40 | traN | 46190..49882 [+], 3693 | Conjugative transfer protein, TraN | TraN_F, T4SS component | |
| 41 | ICEValHN396_041 | 50579..50818 [+], 240 | Hypothetical protein | ||
| 42 | ICEValHN396_042 | 50961..51554 [+], 594 | ATP-binding protein | ||
| 43 | ICEValHN396_043 | 51759..52163 [+], 405 | putative transcriptional regulator | ||
| 44 | ICEValHN396_044 | 52477..54027 [+], 1551 | Aerotaxis sensor receptor protein | ||
| 45 | cheV | 54024..54950 [+], 927 | Chemotaxis protein, CheV | ||
| 46 | ICEValHN396_046 | 54964..56124 [+], 1161 | GGDEF family protein | ||
| 47 | ICEValHN396_047 | 56127..57416 [+], 1290 | Methyl-accepting chemotaxis protein | ||
| 48 | ICEValHN396_048 | 57537..58064 [-], 528 | Transposase | ||
| 49 | s063 | 58115..58702 [-], 588 | Hypothetical protein,S063 | ||
| 50 | s089 | 59070..59414 [+], 345 | Hypothetical protein, S089 | ||
| 51 | ssb | 59411..59830 [+], 420 | Single-stranded DNA-binding protein, Ssb | ||
| 52 | bet | 59910..60728 [+], 819 | Recombination protein, Bet | ||
| 53 | orfZ | 60812..60955 [+], 144 | Hypothetical protein, OrfZ | ||
| 54 | exo | 61016..62032 [+], 1017 | Recombination related exonuclease, Exo | ||
| 55 | cobS | 62242..63201 [+], 960 | Aerobic cobaltochelatase, CobS | ||
| 56 | s088 | 63201..63968 [+], 768 | Hypothetical protein,S088 | ||
| 57 | s068 | 64067..65020 [+], 954 | Cobalamine biosynthesis protein, S068 | ||
| 58 | s069 | 65082..65522 [+], 441 | Hypothetical protein,S069 | ||
| 59 | s070 | 65592..67247 [+], 1656 | Plasmid associated protein, S070 | ||
| 60 | radC | 67331..67828 [+], 498 | DNA repair protein, RadC | ||
| 61 | s092 | 67828..68169 [+], 342 | Hypothetical protein, S092 | ||
| 62 | s072 | 68258..69325 [+], 1068 | Putative primase,S072 | ||
| 63 | s073 | 69414..70121 [+], 708 | Hypothetical protein, S073 | ||
| 64 | ICEValHN396_064 | 70277..70594 [-], 318 | putative Transposase | ||
| 65 | recQ | 71022..73151 [+], 2130 | ATP-dependent DNA helicase, RecQ | ||
| 66 | dprA | 73288..74631 [+], 1344 | DNA recombination-mediator protein A, DprA | ||
| 67 | ICEValHN396_067 | 74678..74812 [-], 135 | Hypothetical protein | ||
| 68 | traF | 75055..75999 [+], 945 | Conjugative transfer pilus assembly protein, TraF | TraF_F, T4SS component | |
| 69 | traH | 76002..77390 [+], 1389 | Conjugative transfer pilus assembly protein, TraH | TraH_F, T4SS component | |
| 70 | traG | 77394..80963 [+], 3570 | Conjugative transfer protein, TraG | TraG_F, T4SS component | |
| 71 | eex | 80994..81449 [-], 456 | Exclusion system protein, Eex | ||
| 72 | ICEValHN396_072 | 81464..82558 [-], 1095 | DDE superfamily endonuclease | ||
| 73 | setC | 82597..83115 [-], 519 | Transcriptional activator, SetC | ||
| 74 | setD | 83108..83410 [-], 303 | Transcriptional activator, SetD | ||
| 75 | ICEValHN396_075 | 83407..83955 [-], 549 | LysM/invasin protein | Orf169_F, T4SS component | |
| 76 | s083 | 83942..84604 [-], 663 | Hypothetical protein, S083 | ||
| 77 | s084 | 84591..85460 [-], 870 | Hypothetical protein, S084 | ||
| 78 | setQ | 85516..85767 [-], 252 | Hypothetical protein, SetQ | ||
| 79 | setR | 85884..86531 [+], 648 | cI prophage repressor protein, setR | ||
| (1) Luo P; He X; Wang Y; Liu Q; Hu C (2016). Comparative genomic analysis of six new-found integrative conjugative elements (ICEs) in Vibrio alginolyticus. BMC Microbiol. 0.721527778. [PubMed:27145747] |