
| 1029 | |
| ICEPmiCHN901 | |
| Proteus mirabilis MD20140901 | |
| 89,493 | |
| 47.3 | |
| - | |
| Resistance to ampicillin, azithromycin, aztreonam, amikacin, chloramphenicol, cefuroxime, cefazolin, ceftriaxone, ciprofloxacin, gentamycin, ampicillin-sulbactam, kanamycin, streptomycin, trimethoprim-sulfamethoxazole, sulfamethoxazole, tetracycline | |
| - | |
| KX243408 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..89493 | |
| coordinates: 3685..3791; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 38804..40954; Gene: traI; Family: MOBH |
The graph information of ICEPmiCHN901 components from KX243408 | |||||
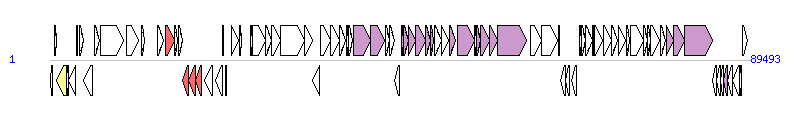 | |||||
| Complete gene list of ICEPmiCHN901 from KX243408 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 127..270 [-], 144 | hypothetical protein | ||
| 2 | - | 615..809 [+], 195 | hypothetical protein | ||
| 3 | int | 826..2067 [-], 1242 | integrase | Integrase | |
| 4 | - | 2069..2338 [-], 270 | hypothetical protein | ||
| 5 | - | 2341..3315 [-], 975 | Rod shape determination protein | ||
| 6 | - | 3390..3578 [+], 189 | hypothetical protein | ||
| 7 | - | 3780..4223 [+], 444 | hypothetical protein | ||
| 8 | rumB | 4234..5406 [-], 1173 | Error-prone, lesion bypass DNA polymerase V (UmuC) | ||
| 9 | tnp | 5690..6283 [+], 594 | Transposase | ||
| 10 | tnpA | 6397..9375 [+], 2979 | Transposase | ||
| 11 | tnpB | 9793..11286 [+], 1494 | Transposase | ||
| 12 | dhfR | 11638..12144 [+], 507 | Dihydrofolate reductase | ||
| 13 | - | 13800..14519 [+], 720 | Type IV secretory pathway, VirD2 components | ||
| 14 | floR | 14736..15950 [+], 1215 | Bicyclomycin resistance protein | AR | |
| 15 | - | 15978..16283 [+], 306 | LysR family transcriptional regulator | ||
| 16 | tnpB | 16395..16934 [+], 540 | Transposase | ||
| 17 | strB | 16906..17736 [-], 831 | streptomycin phosphotransferase | AR | |
| 18 | strA | 17742..18545 [-], 804 | streptomycin phosphotransferase | AR | |
| 19 | sul2 | 18606..19421 [-], 816 | Dihydropteroate synthase | AR | |
| 20 | - | 19699..20748 [-], 1050 | hypothetical protein | ||
| 21 | - | 21134..21997 [-], 864 | Beta-lactamase | ||
| 22 | - | 22028..22159 [+], 132 | hypothetical protein | ||
| 23 | rumA | 22428..22574 [-], 147 | Error-prone repair protein UmuD | ||
| 24 | - | 23216..24121 [+], 906 | DNA polymerase III | ||
| 25 | - | 24209..24508 [+], 300 | hypothetical protein | ||
| 26 | - | 25643..25837 [+], 195 | hypothetical protein | ||
| 27 | - | 25846..27561 [+], 1716 | Type I restriction-modification system, DNA-methyltransferase subunit M | ||
| 28 | - | 27554..28258 [+], 705 | Type I restriction-modification system, specificity subunit S | ||
| 29 | - | 28255..29409 [+], 1155 | hypothetical protein | ||
| 30 | - | 29414..32578 [+], 3165 | Type I restriction-modification system, restriction subunit R | ||
| 31 | - | 32594..33580 [+], 987 | hypothetical protein | ||
| 32 | tnp | 33594..34514 [-], 921 | Mobile element protein | ||
| 33 | - | 34644..35855 [+], 1212 | enzyme | ||
| 34 | - | 35855..36982 [+], 1128 | putative mcrC protein | ||
| 35 | mrr | 36982..37836 [+], 855 | Mrr restriction system protein | ||
| 36 | - | 37930..38211 [+], 282 | hypothetical protein | ||
| 37 | - | 38211..38684 [+], 474 | hypothetical protein | ||
| 38 | traI | 38804..40954 [+], 2151 | Conjugative transfer protein TraI, relaxase | Relaxase, MOBH Family | |
| 39 | traD | 41003..42823 [+], 1821 | IncF plasmid conjugative transfer protein TraD | TraD_F, T4SS component | |
| 40 | - | 42833..43393 [+], 561 | Conjugative transfer protein 234 | ||
| 41 | - | 43380..44015 [+], 636 | Conjugative transfer protein s043 | ||
| 42 | - | 44042..44629 [-], 588 | hypothetical protein | ||
| 43 | traL | 44918..45199 [+], 282 | IncF plasmid conjugative transfer pilus assembly protein TraL | TraL_F, T4SS component | |
| 44 | traE | 45196..45822 [+], 627 | IncF plasmid conjugative transfer pilus assembly protein TraE | TraE_F, T4SS component | |
| 45 | traK | 45806..46702 [+], 897 | IncF plasmid conjugative transfer pilus assembly protein TraK | TraK_F, T4SS component | |
| 46 | traB | 46705..47994 [+], 1290 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F, T4SS component | |
| 47 | traV | 48069..48641 [+], 573 | Conjugative transfer protein TraV | TraV_F, T4SS component | |
| 48 | traA | 48638..49024 [+], 387 | Conjugative transfer protein TraA | TraA_F, T4SS component | |
| 49 | ynd | 49203..50036 [+], 834 | Ynd | ||
| 50 | ync | 50029..50967 [+], 939 | Ync | ||
| 51 | - | 51099..51791 [+], 693 | Thiol:disulfide involved in conjugative transfer | TrbB_I, T4SS component | |
| 52 | traC | 52086..54233 [+], 2148 | IncF plasmid conjugative transfer pilus assembly protein TraC | TraC_F, T4SS component | |
| 53 | - | 54226..54573 [+], 348 | Conjugative transfer protein 345 | ||
| 54 | trhF | 54557..55069 [+], 513 | Conjugative signal peptidase TrhF | TraF, T4SS component | |
| 55 | traW | 55080..56204 [+], 1125 | IncF plasmid conjugative transfer pilus assembly protein TraW | TraW_F, T4SS component | |
| 56 | traU | 56236..57216 [+], 981 | IncF plasmid conjugative transfer pilus assembly protein TraU | TraU_F, T4SS component | |
| 57 | traN | 57219..60911 [+], 3693 | IncF plasmid conjugative transfer protein TraN | TraN_F, T4SS component | |
| 58 | - | 61370..62788 [+], 1419 | hypothetical protein | ||
| 59 | herA | 62785..64824 [+], 2040 | Bipolar DNA helicase HerA | ||
| 60 | - | 65070..65183 [+], 114 | hypothetical protein | ||
| 61 | dns | 65233..65946 [-], 714 | Endonuclease I precursor | ||
| 62 | - | 65949..66401 [-], 453 | Il-IS_2, transposase | ||
| 63 | - | 66672..67274 [-], 603 | hypothetical protein | ||
| 64 | - | 67641..67967 [+], 327 | hypothetical protein | ||
| 65 | ssb | 67983..68402 [+], 420 | Single-stranded DNA-binding protein | ||
| 66 | bet | 68482..69300 [+], 819 | Recombination protein BET | ||
| 67 | - | 69383..69526 [+], 144 | hypothetical protein | ||
| 68 | - | 69587..70603 [+], 1017 | hypothetical protein | ||
| 69 | - | 70801..71772 [+], 972 | Aerobic cobaltochelatase CobS subunit | ||
| 70 | - | 71772..72539 [+], 768 | hypothetical protein | ||
| 71 | - | 72638..73591 [+], 954 | Cobalamine biosynthesis protein | ||
| 72 | - | 73653..74093 [+], 441 | hypothetical protein | ||
| 73 | - | 74163..75818 [+], 1656 | Plasmid associated gene product | ||
| 74 | - | 75901..76398 [+], 498 | DNA repair protein RadC | ||
| 75 | - | 76398..76739 [+], 342 | hypothetical protein | ||
| 76 | - | 76831..77904 [+], 1074 | putative primase | ||
| 77 | - | 77994..78698 [+], 705 | hypothetical protein | ||
| 78 | traF | 78791..79735 [+], 945 | IncF plasmid conjugative transfer pilus assembly protein TraF | TraF_F, T4SS component | |
| 79 | traH | 79738..81126 [+], 1389 | IncF plasmid conjugative transfer pilus assembly protein TraH | TraH_F, T4SS component | |
| 80 | traG | 81130..84699 [+], 3570 | IncF plasmid conjugative transfer protein TraG | TraG_F, T4SS component | |
| 81 | eexS | 84732..85187 [-], 456 | EexS | ||
| 82 | setC | 85221..85754 [-], 534 | Transcriptional activator | ||
| 83 | setD | 85751..86050 [-], 300 | Transcriptional activator | ||
| 84 | - | 86047..86595 [-], 549 | Soluble lytic murein transglycosylase and related regulatory proteins | Orf169_F, T4SS component | |
| 85 | - | 86582..87244 [-], 663 | hypothetical protein | ||
| 86 | - | 87231..88100 [-], 870 | hypothetical protein | ||
| 87 | - | 88156..88407 [-], 252 | hypothetical protein | ||
| 88 | setR | 88525..89172 [+], 648 | Putative cI prophage repressor protein | ||
|
(1) Li X; Du Y; Du P; Dai H; Fang Y; Li Z; Lv N; Zhu B; Kan B; Wang D (2016). SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci Rep. 6:37372. [PubMed:27892525] |