
| 1026 | |
| ICEPmiCHN3300 | |
| Proteus mirabilis TJ3300 | |
| 108,335 | |
| 46.69 | |
| - | |
| Resistance to azithromycin, chloramphenicol, streptomycin, trimethoprim-sulfamethoxazole, sulfamethoxazole | |
| - | |
| KX243415 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..108335 | |
| coordinates: 5813..5919; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 45552..47702; Gene: traI; Family: MOBH |
The graph information of ICEPmiCHN3300 components from KX243415 | |||||
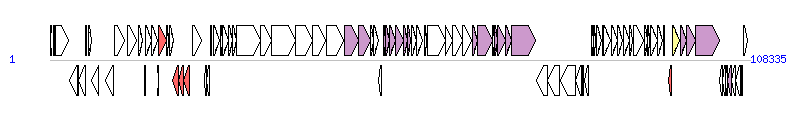 | |||||
| Complete gene list of ICEPmiCHN3300 from KX243415 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 8..136 [+], 129 | hypothetical protein | ||
| 2 | - | 472..660 [+], 189 | hypothetical protein | ||
| 3 | mutL | 806..2824 [+], 2019 | DNA mismatch repair protein MutL | ||
| 4 | int | 2954..4195 [-], 1242 | integrase | ||
| 5 | - | 4197..4466 [-], 270 | hypothetical protein | ||
| 6 | - | 4469..5443 [-], 975 | Rod shape determination protein | ||
| 7 | - | 5539..5706 [+], 168 | hypothetical protein | ||
| 8 | - | 5908..6351 [+], 444 | hypothetical protein | ||
| 9 | rumB | 6362..7534 [-], 1173 | Error-prone, lesion bypass DNA polymerase V (UmuC) | ||
| 10 | tnp | 8664..9842 [-], 1179 | Transposase | ||
| 11 | tnpA | 10028..11503 [+], 1476 | Transposase | ||
| 12 | tnpB | 11921..13414 [+], 1494 | Transposase | ||
| 13 | dhfR | 13766..14272 [+], 507 | Dihydrofolate reductase | ||
| 14 | - | 14650..14808 [-], 159 | hypothetical protein | ||
| 15 | tnp | 14809..15732 [+], 924 | Mobile element protein | ||
| 16 | - | 15763..16647 [+], 885 | Type IV secretory pathway, VirD2 components | ||
| 17 | - | 16743..16865 [-], 123 | hypothetical protein | ||
| 18 | floR | 16864..18078 [+], 1215 | Bicyclomycin resistance protein | AR | |
| 19 | - | 18106..18411 [+], 306 | Transcriptional regulator | ||
| 20 | tnpB | 18523..19062 [+], 540 | Mobile element protein | ||
| 21 | strB | 19034..19870 [-], 837 | streptomycin phosphotransferase | AR | |
| 22 | strA | 19870..20673 [-], 804 | streptomycin phosphotransferase | AR | |
| 23 | sul2 | 20734..21549 [-], 816 | Dihydropteroate synthase | AR | |
| 24 | tnpA | 22020..23393 [+], 1374 | Transposase | ||
| 25 | rumB | 23978..24247 [-], 270 | Error-prone, lesion bypass DNA polymerase V (UmuC) | ||
| 26 | rumA | 24255..24704 [-], 450 | Error-prone repair protein UmuD | ||
| 27 | - | 24823..24960 [+], 138 | hypothetical protein | ||
| 28 | - | 25343..26248 [+], 906 | DNA polymerase III | ||
| 29 | - | 26447..26650 [+], 204 | hypothetical protein | ||
| 30 | - | 26758..27645 [+], 888 | hypothetical protein | ||
| 31 | - | 27655..28254 [+], 600 | putative inner membrane protein | ||
| 32 | - | 28251..28835 [+], 585 | putative cytoplasmic protein | ||
| 33 | - | 28854..32531 [+], 3678 | hypothetical protein | ||
| 34 | - | 32548..34311 [+], 1764 | hypothetical protein | ||
| 35 | - | 34327..37953 [+], 3627 | putative type II restriction enzyme methylase subunit | ||
| 36 | - | 37953..40610 [+], 2658 | putative cytoplasmic protein | ||
| 37 | - | 40627..42702 [+], 2076 | ATP-dependent protease Type I | ||
| 38 | - | 42746..45397 [+], 2652 | ATPase involved in DNA repair | ||
| 39 | traI | 45552..47702 [+], 2151 | Conjugative transfer protein TraI, relaxase | Relaxase, MOBH Family | |
| 40 | traD | 47751..49571 [+], 1821 | IncF plasmid conjugative transfer protein TraD | TraD_F, T4SS component | |
| 41 | - | 49581..50141 [+], 561 | Conjugative transfer protein 234 | ||
| 42 | - | 50128..50763 [+], 636 | Conjugative transfer protein s043 | ||
| 43 | - | 50790..51377 [-], 588 | hypothetical protein | ||
| 44 | traL | 51666..51947 [+], 282 | IncF plasmid conjugative transfer pilus assembly protein TraL | TraL_F, T4SS component | |
| 45 | traE | 51944..52570 [+], 627 | IncF plasmid conjugative transfer pilus assembly protein TraE | TraE_F, T4SS component | |
| 46 | traK | 52554..53453 [+], 900 | IncF plasmid conjugative transfer pilus assembly protein TraK | TraK_F, T4SS component | |
| 47 | traB | 53453..54742 [+], 1290 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F, T4SS component | |
| 48 | traV | 54739..55389 [+], 651 | Conjugative transfer protein TraV | TraV_F, T4SS component | |
| 49 | traA | 55386..55772 [+], 387 | Conjugative transfer protein TraA | TraA_F, T4SS component | |
| 50 | ynd | 55950..56783 [+], 834 | Ynd | ||
| 51 | ync | 56776..57714 [+], 939 | Ync | ||
| 52 | - | 58023..58196 [+], 174 | hypothetical protein | ||
| 53 | - | 58423..61152 [+], 2730 | putative ATP-dependent helicase | ||
| 54 | - | 61149..62258 [+], 1110 | hypothetical protein | ||
| 55 | - | 62271..63803 [+], 1533 | hypothetical protein | ||
| 56 | - | 63806..65251 [+], 1446 | Response regulator of zinc sigma-54-dependent two-component system | ||
| 57 | - | 65396..66088 [+], 693 | Thiol:disulfide involved in conjugative transfer | TrbB_I, T4SS component | |
| 58 | traC | 66088..68487 [+], 2400 | IncF plasmid conjugative transfer pilus assembly protein TraC | TraC_F, T4SS component | |
| 59 | - | 68480..68827 [+], 348 | Conjugative transfer protein 345 | ||
| 60 | trhF | 68811..69323 [+], 513 | Conjugative signal peptidase TrhF | TraF, T4SS component | |
| 61 | traW | 69334..70458 [+], 1125 | IncF plasmid conjugative transfer pilus assembly protein TraW | TraW_F, T4SS component | |
| 62 | traU | 70442..71470 [+], 1029 | IncF plasmid conjugative transfer pilus assembly protein TraU | TraU_F, T4SS component | |
| 63 | traN | 71473..75165 [+], 3693 | IncF plasmid conjugative transfer protein TraN | TraN_F, T4SS component | |
| 64 | prcA | 75288..76964 [-], 1677 | ATP-dependent DNA helicase pcrA | ||
| 65 | - | 76961..78886 [-], 1926 | putative exonuclease | ||
| 66 | - | 78964..81342 [-], 2379 | putative serine protease | ||
| 67 | - | 81358..82329 [-], 972 | Cell division protein FtsH | ||
| 68 | - | 82440..82619 [-], 180 | hypothetical protein | ||
| 69 | - | 82761..83363 [-], 603 | hypothetical protein | ||
| 70 | - | 83773..84057 [+], 285 | hypothetical protein | ||
| 71 | ssb | 84073..84492 [+], 420 | Single-stranded DNA-binding protein | ||
| 72 | bet | 84572..85390 [+], 819 | Recombination protein BET | ||
| 73 | - | 85473..85616 [+], 144 | hypothetical protein | ||
| 74 | - | 85677..86693 [+], 1017 | hypothetical protein | ||
| 75 | - | 86903..87862 [+], 960 | Aerobic cobaltochelatase CobS subunit | ||
| 76 | - | 87865..88629 [+], 765 | hypothetical protein | ||
| 77 | - | 88728..89681 [+], 954 | Cobalamine biosynthesis protein | ||
| 78 | - | 89743..90183 [+], 441 | hypothetical protein | ||
| 79 | - | 90253..91908 [+], 1656 | Plasmid associated gene product | ||
| 80 | - | 91992..92489 [+], 498 | DNA repair protein RadC | ||
| 81 | - | 92489..92830 [+], 342 | hypothetical protein | ||
| 82 | - | 92922..93995 [+], 1074 | putative primase | ||
| 83 | - | 94084..94791 [+], 708 | hypothetical protein | ||
| 84 | - | 95022..95162 [+], 141 | hypothetical protein | ||
| 85 | dhfR | 95651..96124 [-], 474 | Dihydrofolate reductase | AR | |
| 86 | - | 96412..97371 [+], 960 | intIPac | Integrase | |
| 87 | traF | 97633..98577 [+], 945 | IncF plasmid conjugative transfer pilus assembly protein TraF | TraF_F, T4SS component | |
| 88 | traH | 98580..99968 [+], 1389 | IncF plasmid conjugative transfer pilus assembly protein TraH | TraH_F, T4SS component | |
| 89 | traG | 99972..103541 [+], 3570 | IncF plasmid conjugative transfer protein TraG | TraG_F, T4SS component | |
| 90 | eexR | 103574..104005 [-], 432 | EexR | ||
| 91 | setC | 104063..104596 [-], 534 | Transcriptional activator | ||
| 92 | setD | 104593..104892 [-], 300 | Transcriptional activator | ||
| 93 | - | 104889..105437 [-], 549 | Soluble lytic murein transglycosylase and related regulatory proteins (some containing LysM/invasin domains) | Orf169_F, T4SS component | |
| 94 | - | 105424..106086 [-], 663 | hypothetical protein | ||
| 95 | - | 106073..106942 [-], 870 | hypothetical protein | ||
| 96 | - | 106998..107249 [-], 252 | hypothetical protein | ||
| 97 | setR | 107367..108014 [+], 648 | Putative cI prophage repressor protein | ||
|
(1) Li X; Du Y; Du P; Dai H; Fang Y; Li Z; Lv N; Zhu B; Kan B; Wang D (2016). SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci Rep. 6:37372. [PubMed:27892525] |