
| 1019 | |
| ICEPmiChn2 | |
| ICEO_0000148 | |
| Proteus mirabilis JN7 | |
| 104,371 | |
| 47.34 | |
| - | |
| Antibiotic resistance genes: floR, sul2 | |
| Escherichia coli J53 | |
| KY437726 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..104371 | |
| coordinates: 3980..4086; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 43070..45220; Gene: traI; Family: MOBH |
The graph information of ICEPmiChn2 components from KY437726 | |||||
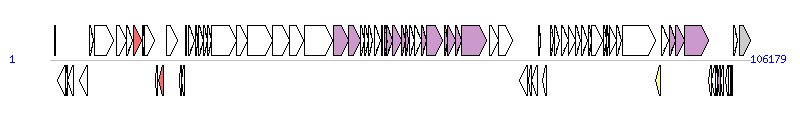 | |||||
| Complete gene list of ICEPmiChn2 from KY437726 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | xis | 630..791 [+], 162 | Xis | ||
| 2 | int | 1121..2362 [-], 1242 | integrase | ||
| 3 | - | 2364..2633 [-], 270 | unknown | ||
| 4 | - | 2636..3610 [-], 975 | hypothetical protein | ||
| 5 | rumB | 4529..5701 [-], 1173 | UV repair DNA polymerase | ||
| 6 | tnp | 5985..6578 [+], 594 | putative transposase | ||
| 7 | tnpA | 6692..9670 [+], 2979 | transposase | ||
| 8 | tnpB | 10088..11581 [+], 1494 | putative transposase | ||
| 9 | - | 11612..12496 [+], 885 | hypothetical protein | ||
| 10 | floR | 12713..13927 [+], 1215 | florfenicol exporter | AR | |
| 11 | lysR | 13955..14260 [+], 306 | hypothetical protein | ||
| 12 | tnpA | 14372..15865 [+], 1494 | transposase | ||
| 13 | glmM | 16041..16343 [-], 303 | phosphoglucosamine mutase | ||
| 14 | sul2 | 16430..17245 [-], 816 | sulfonamide-resistance dihydropteroate synthase protein Sul2 | AR | |
| 15 | tnpA | 17716..19329 [+], 1614 | transposase | ||
| 16 | - | 19672..19941 [-], 270 | ultraviolet light resistance protein B | ||
| 17 | - | 19949..20398 [-], 450 | protein UmuD | ||
| 18 | - | 20517..20654 [+], 138 | hypothetical protein | ||
| 19 | - | 21037..21942 [+], 906 | exonuclease, RNase T and DNA polymerase III | ||
| 20 | - | 22030..22344 [+], 315 | conserved hypothetical protein | ||
| 21 | - | 22452..23339 [+], 888 | conserved hypothetical protein | ||
| 22 | - | 23349..23948 [+], 600 | conserved hypothetical protein | ||
| 23 | - | 23945..24529 [+], 585 | conserved hypothetical protein | ||
| 24 | - | 24548..28225 [+], 3678 | conserved hypothetical protein | ||
| 25 | chn2-1 | 28242..30005 [+], 1764 | hypothetical protein | ||
| 26 | chn2-2 | 30021..33722 [+], 3702 | putative restriction enzyme; adenosylmethionine-dependent methyltransferases | ||
| 27 | chn2-3 | 33722..36379 [+], 2658 | putative alkaline phosphatase protein | ||
| 28 | chn2-4 | 36396..38471 [+], 2076 | putative ATP-dependent Lon protease; turnover of interacellular proteins function | ||
| 29 | chn2-5 | 38622..42914 [+], 4293 | hypothetical protein | ||
| 30 | traI | 43070..45220 [+], 2151 | TraI | Relaxase, MOBH Family | |
| 31 | traD | 45269..47089 [+], 1821 | TraD | TraD_F, T4SS component | |
| 32 | - | 47099..47659 [+], 561 | hypothetical protein | ||
| 33 | - | 47646..48281 [+], 636 | conjugative transfer protein s043 | ||
| 34 | enispa1-1 | 48353..48979 [+], 627 | hypothetical protein | ||
| 35 | enispa1-2 | 49192..50091 [+], 900 | putative Zn peptidase | ||
| 36 | traL | 50228..50509 [+], 282 | TraL | TraL_F, T4SS component | |
| 37 | traE | 50740..51132 [+], 393 | TraE | TraE_F, T4SS component | |
| 38 | traK | 51113..52012 [+], 900 | TraK | TraK_F, T4SS component | |
| 39 | traB | 52015..53304 [+], 1290 | TraB | TraB_F, T4SS component | |
| 40 | htdD | 53301..53951 [+], 651 | HtdD | TraV_F, T4SS component | |
| 41 | traA | 53948..54334 [+], 387 | TraA | TraA_F, T4SS component | |
| 42 | ynd | 54513..55346 [+], 834 | Ynd | ||
| 43 | ync | 55339..56277 [+], 939 | Ync | ||
| 44 | dsbC | 56409..57101 [+], 693 | thiol:disulfide interchange protein DsbC | TrbB_I, T4SS component | |
| 45 | traC | 57101..59500 [+], 2400 | TraC | TraC_F, T4SS component | |
| 46 | trbI | 59824..60336 [+], 513 | TrbI | TraF, T4SS component | |
| 47 | traW | 60347..61471 [+], 1125 | TraW | TraW_F, T4SS component | |
| 48 | traU | 61455..62483 [+], 1029 | TraU | TraU_F, T4SS component | |
| 49 | traN | 62486..66178 [+], 3693 | TraN | TraN_F, T4SS component | |
| 50 | chn1 | 66637..68055 [+], 1419 | hypothetical protein | ||
| 51 | chn2 | 68052..70091 [+], 2040 | hypothetical protein | ||
| 52 | ISPpu12 tnpA | 71216..72430 [-], 1215 | putative transposase TnpA | ||
| 53 | lspA | 72529..73041 [-], 513 | putative lipoprotein signal peptidase LspA | ||
| 54 | orf1 | 73045..73941 [-], 897 | putative cation-efflux family protein | ||
| 55 | orf2 | 74037..74444 [+], 408 | putative MerR family regulator | ||
| 56 | - | 74674..75276 [-], 603 | hypothetical protein | ||
| 57 | ssb | 75986..76405 [+], 420 | single-stranded DNA binding protein | ||
| 58 | bet | 76485..77303 [+], 819 | recombination protein, Bet | ||
| 59 | exo | 77591..78607 [+], 1017 | recombination related exonuclease, Exo | ||
| 60 | cobS | 78700..79776 [+], 1077 | aerobic cobaltochelatase CobS subunit | ||
| 61 | - | 79776..80543 [+], 768 | hypothetical protein | ||
| 62 | - | 80642..81595 [+], 954 | hypothetical protein | ||
| 63 | - | 81657..82097 [+], 441 | hypothetical protein | ||
| 64 | - | 82167..83822 [+], 1656 | hypothetical protein | ||
| 65 | - | 83906..84403 [+], 498 | DNA repair protein RadC | ||
| 66 | - | 84403..84744 [+], 342 | hypothetical protein | ||
| 67 | - | 84836..85909 [+], 1074 | putative primase | ||
| 68 | - | 85999..86703 [+], 705 | hypothetical protein | ||
| 69 | chn2-6 | 86904..91820 [+], 4917 | unknown | ||
| 70 | chn2-7 | 91783..92634 [-], 852 | unknown | Integrase | |
| 71 | chn2-8 | 92827..93825 [+], 999 | unknown | ||
| 72 | traF | 93964..94896 [+], 933 | TraF | TraF_F, T4SS component | |
| 73 | traH | 94899..96287 [+], 1389 | TraH | TraH_F, T4SS component | |
| 74 | traG | 96291..99860 [+], 3570 | TraG | TraG_F, T4SS component | |
| 75 | eexS | 99893..100348 [-], 456 | EexS | ||
| 76 | setC | 100382..100915 [-], 534 | transcriptional activator | ||
| 77 | setD | 100912..101211 [-], 300 | transcriptional activator | ||
| 78 | - | 101208..101756 [-], 549 | hypothetical protein | Orf169_F, T4SS component | |
| 79 | - | 101743..102111 [-], 369 | hypothetical protein | ||
| 80 | - | 102392..103261 [-], 870 | hypothetical protein | ||
| 81 | - | 102392..102913 [-], 522 | hypothetical protein | ||
| 82 | - | 103317..103568 [-], 252 | hypothetical protein | ||
| 83 | setR | 103686..104333 [+], 648 | transcriptional repressor | ||
| 84 | prfC | 104590..106179 [+], 1590 | peptide chain release factor 3 | ||
|
(1) Bie L; Wu H; Wang XH; Wang M; Xu H (2017). Identification and characterization of new members of the SXT/R391 family of integrative and conjugative elements (ICEs) in Proteus mirabilis. Int J Antimicrob Agents. 50(2):242-246. [PubMed:28602701] |