
| 1018 | |
| ICEPmiCHN1809 | |
| ICEO_0000149 | |
| Proteus mirabilis TJ1809 | |
| 76,218 | |
| 47.11 | |
| - | |
| Resistance to ampicillin, azithromycin, chloramphenicol, ciprofloxacin, kanamycin, trimethoprim-sulfamethoxazole, sulfamethoxazole, tetracycline | |
| - | |
| KX243413 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..76218 | |
| coordinates: 4064..4170; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTTACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTTGGGGTTAATTGGATGGGGAA | |
| coordinates: 21095..23245; Gene: traI; Family: MOBH |
The graph information of ICEPmiCHN1809 components from KX243413 | |||||
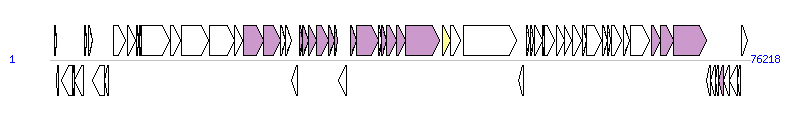 | |||||
| Complete gene list of ICEPmiCHN1809 from KX243413 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | - | 472..666 [+], 195 | hypothetical protein | ||
| 2 | - | 718..930 [-], 213 | hypothetical protein | ||
| 3 | int | 1205..2446 [-], 1242 | integrase | ||
| 4 | - | 2448..2717 [-], 270 | hypothetical protein | ||
| 5 | - | 2720..3694 [-], 975 | Rod shape determination protein | ||
| 6 | - | 3790..3957 [+], 168 | hypothetical protein | ||
| 7 | - | 4159..4602 [+], 444 | hypothetical protein | ||
| 8 | rumB | 4613..5881 [-], 1269 | Error-prone, lesion bypass DNA polymerase V (UmuC) | ||
| 9 | rumA | 5889..6338 [-], 450 | Error-prone repair protein UmuD | ||
| 10 | - | 6936..8204 [+], 1269 | transposase,IS4 | ||
| 11 | - | 8387..9295 [+], 909 | DNA polymerase III | ||
| 12 | - | 9369..9668 [+], 300 | hypothetical protein | ||
| 13 | - | 9740..9949 [+], 210 | hypothetical protein | ||
| 14 | - | 9933..13058 [+], 3126 | hypothetical protein | ||
| 15 | - | 13104..14186 [+], 1083 | hypothetical protein | ||
| 16 | - | 14306..17308 [+], 3003 | Adenine-specific DNA methylase containing a Zn-ribbon | ||
| 17 | - | 17384..20134 [+], 2751 | putative DEAH ATP-dependent helicase | ||
| 18 | - | 20131..21000 [+], 870 | Mrr restriction endonuclease | ||
| 19 | traI | 21095..23245 [+], 2151 | Conjugative transfer protein TraI, relaxase | Relaxase, MOBH Family | |
| 20 | traD | 23294..25114 [+], 1821 | IncF plasmid conjugative transfer protein TraD | TraD_F, T4SS component | |
| 21 | - | 25124..25684 [+], 561 | Conjugative transfer protein 234 | ||
| 22 | - | 25671..26306 [+], 636 | Conjugative transfer protein s043 | ||
| 23 | - | 26333..26920 [-], 588 | hypothetical protein | ||
| 24 | traL | 27209..27490 [+], 282 | IncF plasmid conjugative transfer pilus assembly protein TraL | TraL_F, T4SS component | |
| 25 | traE | 27487..28113 [+], 627 | IncF plasmid conjugative transfer pilus assembly protein TraE | TraE_F, T4SS component | |
| 26 | traK | 28097..28993 [+], 897 | IncF plasmid conjugative transfer pilus assembly protein TraK | TraK_F, T4SS component | |
| 27 | traB | 28996..30285 [+], 1290 | IncF plasmid conjugative transfer pilus assembly protein TraB | TraB_F, T4SS component | |
| 28 | traV | 30360..30932 [+], 573 | Conjugative transfer protein TraV | TraV_F, T4SS component | |
| 29 | traA | 30929..31315 [+], 387 | Conjugative transfer protein TraA | TraA_F, T4SS component | |
| 30 | - | 31396..32334 [-], 939 | TIORF34 protein | ||
| 31 | - | 32676..33368 [+], 693 | Thiol:disulfide involved in conjugative transfer | TrbB_I, T4SS component | |
| 32 | traC | 33368..35767 [+], 2400 | IncF plasmid conjugative transfer pilus assembly protein TraC | TraC_F, T4SS component | |
| 33 | - | 35760..36107 [+], 348 | Conjugative transfer protein 345 | ||
| 34 | trhF | 36091..36603 [+], 513 | Conjugative signal peptidase TrhF | TraF, T4SS component | |
| 35 | traW | 36614..37738 [+], 1125 | IncF plasmid conjugative transfer pilus assembly protein TraW | TraW_F, T4SS component | |
| 36 | traU | 37770..38750 [+], 981 | IncF plasmid conjugative transfer pilus assembly protein TraU | TraU_F, T4SS component | |
| 37 | traN | 38753..42445 [+], 3693 | IncF plasmid conjugative transfer protein TraN | TraN_F, T4SS component | |
| 38 | - | 42755..43645 [+], 891 | putative integrase | Integrase | |
| 39 | - | 43642..44694 [+], 1053 | transposase | ||
| 40 | - | 45061..50835 [+], 5775 | serine protease-like | ||
| 41 | - | 50959..51561 [-], 603 | hypothetical protein | ||
| 42 | - | 51929..52255 [+], 327 | hypothetical protein | ||
| 43 | ssb | 52271..52690 [+], 420 | Single-stranded DNA-binding protein | ||
| 44 | bet | 52770..53588 [+], 819 | Recombination protein BET | ||
| 45 | - | 53671..53814 [+], 144 | hypothetical protein | ||
| 46 | - | 53875..54891 [+], 1017 | hypothetical protein | ||
| 47 | - | 55101..56060 [+], 960 | Aerobic cobaltochelatase CobS subunit | ||
| 48 | - | 56060..56827 [+], 768 | hypothetical protein | ||
| 49 | - | 56926..57879 [+], 954 | Cobalamine biosynthesis protein | ||
| 50 | - | 57941..58381 [+], 441 | hypothetical protein | ||
| 51 | - | 58451..60100 [+], 1650 | Plasmid associated gene product | ||
| 52 | - | 60184..60681 [+], 498 | DNA repair protein RadC | ||
| 53 | - | 60681..61022 [+], 342 | hypothetical protein | ||
| 54 | - | 61114..62187 [+], 1074 | putative primase | ||
| 55 | - | 62423..63130 [+], 708 | hypothetical protein | ||
| 56 | - | 63183..65291 [+], 2109 | Translation-disabling ACNase RloC | ||
| 57 | traF | 65521..66465 [+], 945 | IncF plasmid conjugative transfer pilus assembly protein TraF | TraF_F, T4SS component | |
| 58 | traH | 66468..67856 [+], 1389 | IncF plasmid conjugative transfer pilus assembly protein TraH | TraH_F, T4SS component | |
| 59 | traG | 67860..71429 [+], 3570 | IncF plasmid conjugative transfer protein TraG | TraG_F, T4SS component | |
| 60 | eexR | 71460..71891 [-], 432 | EexR | ||
| 61 | setC | 71946..72479 [-], 534 | Transcriptional activator | ||
| 62 | setD | 72476..72775 [-], 300 | Transcriptional activator | ||
| 63 | - | 72772..73320 [-], 549 | Soluble lytic murein transglycosylase and related regulatory proteins | Orf169_F, T4SS component | |
| 64 | - | 73307..73969 [-], 663 | hypothetical protein | ||
| 65 | - | 73956..74825 [-], 870 | hypothetical protein | ||
| 66 | - | 74881..75132 [-], 252 | hypothetical protein | ||
| 67 | setR | 75250..75897 [+], 648 | Putative cI prophage repressor protein | ||
|
(1) Li X; Du Y; Du P; Dai H; Fang Y; Li Z; Lv N; Zhu B; Kan B; Wang D (2016). SXT/R391 integrative and conjugative elements in Proteus species reveal abundant genetic diversity and multidrug resistance. Sci Rep. 6:37372. [PubMed:27892525] |