
| 1016 | |
| ICEPmiChn1 | |
| ICEO_0000018 | |
| Proteus mirabilis | |
| 92751 | |
| 48.01 | |
| prfC | |
| Antibiotic resistance genes: floR, tet(G), strAB and sul2; doxycycline, streptomycin, spectinomycin, trimethoprim-sulfamethoxazole, florfenicol resistance | |
| Escherichia coli | |
| KT962845 (complete ICE sequence in this GenBank file) | |
| - | |
| 1..92751 | |
| coordinates: 3990..4096; oriTDB id: 200056 TATCGAGACGCCAAACAGTGATTGTGACGGCAGTTTTACGTTTGGCGTTTCGATCCAAAAGCCAAACGGA TAGTGGTTTTGGCTTTAGGGGTTAATTGGATGGGGAA | |
| coordinates: 38622..40772; Gene: traI; Family: MOBH |
The graph information of ICEPmiChn1 components from KT962845 | |||||
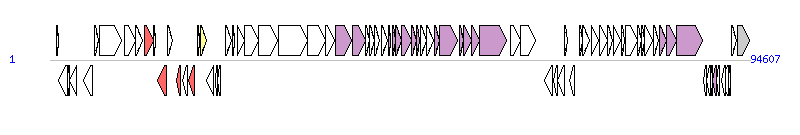 | |||||
| Complete gene list of ICEPmiChn1 from KT962845 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | *Reannotation | |
| 1 | xis | 914..1114 [+], 201 | Xis | ||
| 2 | int | 1131..2372 [-], 1242 | integrase | ||
| 3 | - | 2374..2643 [-], 270 | S002 | ||
| 4 | - | 2646..3620 [-], 975 | hypothetical protein | ||
| 5 | rumB | 4539..5711 [-], 1173 | UV repair DNA polymerase | ||
| 6 | tnp | 5995..6588 [+], 594 | putative transposase | ||
| 7 | tnpA | 6702..9680 [+], 2979 | transposase | ||
| 8 | tnpB | 10098..11591 [+], 1494 | putative transposase | ||
| 9 | - | 11622..12506 [+], 885 | hypothetical protein | ||
| 10 | floR | 12723..13937 [+], 1215 | florfenicol exporter | AR | |
| 11 | lysR | 13965..14270 [+], 306 | hypothetical protein | ||
| 12 | tetA | 14537..15811 [-], 1275 | tetracycline repressor protein TetA | AR | |
| 13 | tetR | 15815..16492 [+], 678 | tetracycline resistance repressor protein TetR | ||
| 14 | strB | 17072..17677 [-], 606 | streptomycin phosphotransferase protein StrB | AR | |
| 15 | strA | 17908..18525 [-], 618 | streptomycin phosphotransferase protein StrA | ||
| 16 | sul2 | 18772..19587 [-], 816 | sulfonamide-resistance dihydropteroate synthase protein Sul2 | AR | |
| 17 | tnpI | 19960..20250 [+], 291 | TnpI transposase | ||
| 18 | tnpII | 20382..21113 [+], 732 | TnpII transposase | Integrase | |
| 19 | - | 21124..22101 [-], 978 | hypothetical protein | ||
| 20 | rumB | 22308..22616 [-], 309 | RumB | ||
| 21 | rumA | 22585..23034 [-], 450 | hypothetical protein | ||
| 22 | - | 23676..24581 [+], 906 | hypothetical protein | ||
| 23 | - | 24669..24968 [+], 300 | hypothetical protein | ||
| 24 | - | 25286..26239 [+], 954 | hypothetical protein | ||
| 25 | - | 26320..28155 [+], 1836 | RM system Met subunit | ||
| 26 | - | 28157..30808 [+], 2652 | RM system Res subunit | ||
| 27 | - | 30905..34786 [+], 3882 | hypothetical protein | ||
| 28 | - | 34741..37212 [+], 2472 | hypothetical protein | ||
| 29 | - | 37284..38465 [+], 1182 | hypothetical protein | ||
| 30 | traI | 38622..40772 [+], 2151 | TraI | Relaxase, MOBH Family | |
| 31 | traD | 40821..42641 [+], 1821 | TraD | TraD_F, T4SS component | |
| 32 | - | 42651..43211 [+], 561 | hypothetical protein | ||
| 33 | - | 43198..43833 [+], 636 | Conjugative transfer protein s043 | ||
| 34 | enispa1-1 | 43905..44531 [+], 627 | hypothetical protein | ||
| 35 | enispa1-2 | 44743..45642 [+], 900 | putative Zn peptidase | ||
| 36 | traL | 45779..46060 [+], 282 | TraL | TraL_F, T4SS component | |
| 37 | traE | 46291..46683 [+], 393 | TraE | TraE_F, T4SS component | |
| 38 | traK | 46664..47563 [+], 900 | TraK | TraK_F, T4SS component | |
| 39 | traB | 47566..48855 [+], 1290 | TraB | TraB_F, T4SS component | |
| 40 | htdD | 48852..49502 [+], 651 | HtdD | TraV_F, T4SS component | |
| 41 | traA | 49499..49885 [+], 387 | TraA | TraA_F, T4SS component | |
| 42 | ynd | 50064..50897 [+], 834 | Ynd | ||
| 43 | ync | 50890..51828 [+], 939 | Ync | ||
| 44 | dsbC | 51960..52652 [+], 693 | thiol:disulfide interchange protein DsbC | TrbB_I, T4SS component | |
| 45 | traC | 52652..55051 [+], 2400 | TraC | TraC_F, T4SS component | |
| 46 | trbI | 55375..55887 [+], 513 | TrbI | TraF, T4SS component | |
| 47 | traW | 55898..57022 [+], 1125 | TraW | TraW_F, T4SS component | |
| 48 | traU | 57006..58034 [+], 1029 | TraU | TraU_F, T4SS component | |
| 49 | traN | 58037..61729 [+], 3693 | TraN | TraN_F, T4SS component | |
| 50 | - | 62188..63606 [+], 1419 | hypothetical protein | ||
| 51 | - | 63603..65642 [+], 2040 | hypothetical protein | ||
| 52 | ISPpu12 tnpA | 66767..67981 [-], 1215 | putative transposase TnpA | ||
| 53 | lspA | 68078..68590 [-], 513 | putative lipoprotein signal peptidase LspA | ||
| 54 | cation-efflux family gene | 68594..69490 [-], 897 | putative cation-efflux family protein | ||
| 55 | putative MerR family gene | 69586..69993 [+], 408 | putative MerR family regulator | ||
| 56 | - | 70223..70825 [-], 603 | Hypothetical protein | ||
| 57 | ssb | 71534..71953 [+], 420 | single-stranded DNA binding protein | ||
| 58 | bet | 72033..72851 [+], 819 | Recombination protein, Bet | ||
| 59 | exo | 73138..74154 [+], 1017 | Recombination related exonuclease, Exo | ||
| 60 | cobS | 74247..75323 [+], 1077 | Aerobic cobaltochelatase CobS subunit | ||
| 61 | - | 75323..76090 [+], 768 | hypothetical protein | ||
| 62 | - | 76189..77142 [+], 954 | hypothetical protein | ||
| 63 | - | 77204..77644 [+], 441 | hypothetical protein | ||
| 64 | - | 77714..79369 [+], 1656 | hypothetical protein | ||
| 65 | - | 79452..79949 [+], 498 | DNA repair protein RadC | ||
| 66 | - | 79949..80290 [+], 342 | hypothetical protein | ||
| 67 | - | 80382..81455 [+], 1074 | putative primase | ||
| 68 | - | 81545..82249 [+], 705 | hypothetical protein | ||
| 69 | traF | 82354..83286 [+], 933 | TraF | TraF_F, T4SS component | |
| 70 | traH | 83289..84677 [+], 1389 | TraH | TraH_F, T4SS component | |
| 71 | traG | 84681..88250 [+], 3570 | TraG | TraG_F, T4SS component | |
| 72 | eexS | 88283..88738 [-], 456 | EexS | ||
| 73 | setC | 88772..89305 [-], 534 | transcriptional activator | ||
| 74 | setD | 89302..89601 [-], 300 | transcriptional activator | ||
| 75 | - | 89598..90146 [-], 549 | hypothetical protein | Orf169_F, T4SS component | |
| 76 | - | 90133..90501 [-], 369 | hypothetical protein | ||
| 77 | - | 90782..91651 [-], 870 | hypothetical protein | ||
| 78 | - | 90782..91303 [-], 522 | hypothetical protein | ||
| 79 | - | 91707..91958 [-], 252 | hypothetical protein | ||
| 80 | setR | 92076..92723 [+], 648 | transcriptional repressor | ||
| 81 | prfC | 92980..94569 [+], 1590 | peptide chain release factor 3 | ||
| (1) Ryan MP; Armshaw P; O'Halloran JA; Pembroke JT (2017). Analysis and comparative genomics of R997, the first SXT/R391 integrative and conjugative element (ICE) of the Indian Sub-Continent. Sci Rep. 7(1):8562. [PubMed:28819148] |
|
(2) Badhai J; Das SK (2016). Characterization of Three Novel SXT/R391 Integrating Conjugative Elements ICEMfuInd1a and ICEMfuInd1b, and ICEMprChn1 Identified in the Genomes of Marinomonas fungiae JCM 18476(T) and Marinomonas profundimaris Strain D104. Front Microbiol. 1.608333333. [PubMed:27933056] |
|
(3) Lei CW; Zhang AY; Wang HN; Liu BH; Yang LQ; Yang YQ (2016). Characterization of SXT/R391 Integrative and Conjugative Elements in Proteus mirabilis Isolates from Food-Producing Animals in China. Antimicrob Agents Chemother. 60(3):1935-8. [PubMed:26824957] |