
| TADB2: a web-based resource for toxin-antitoxin loci in Bacteria and Archaea |
|||||||||||||||||||||||||||||||||||||||||||||||||
Reference: Y. Shao, E.M. Harrison, D. Bi, C. Tai, X. He, H.Y. Ou, K. Rajakumar and Z. Deng (2010) TADB: a web-based resource for Type 2 toxin-antitoxin loci in Bacteria and Archaea. Nucleic Acids Research, doi:10.1093/nar/gkq908. [Abstract] |
|||||||||||||||||||||||||||||||||||||||||||||||||
| TADB2: a web-based resource for toxin-antitoxin loci in Bacteria and Archaea |
|||||||||||||||||||||||||||||||||||||||||||||||||
| TA loci in prokaryote | |||||||||||||||||||||||||||||||||||||||||||||||||
|
Eubacterial and archaeal toxin-antitoxin (TA) loci, typically containing two but occasionally three tandem genes,
code for cognate partners that have been hypothesized or demonstrated to play key roles in stress response,
bacterial physiology and stabilization of horizontally acquired genetic elements.
The toxin genes invariably code for proteins,
while matching antitoxin genes code for either antisense RNA or antitoxin
proteins, resulting in classification as Type 1 or Type 2 TA loci, respectively.
To date numerous phylogenetically and functionally distinct Type 2 TA systems
have been identified through experimental and bioinformatics studies. In
particular, since the advent of the genomics era it has become abundantly
clear that Type 2 TA loci are richly distributed throughout the genomes
of almost all free-living Eubacteria and Archaea . Individual bacterial
and archael genomes may harbour between 0 ~ >50 TA loci, with the vast
majority encoding at least one TA locus. Consistent with the focus of this
study, all further references within this report to 'TA' loci/genes/proteins
refer solely to corresponding counterparts in 'Type 2 TA' systems. TA toxin genes code for stable toxins that target either DNA replication or translation and these toxins are neutralized by short-lived protein antitoxins.As a general model for TA systems, the short lived nature of the antitoxin ensures that cells that lose a TA locus and thus no longer produce the cognate 'immunity' antitoxin protein become susceptible to the stable toxin that has outlived the presence of its coding gene, resulting in toxin-mediated killing of the cell. Hence, prokaryotic organisms harboring TA loci are considered by some investigators to be 'addicted' to their native repertoire of TA loci. TA systems may well contribute towards stochastic variation within the population, gearing up individual subsets of organisms distinctly to favor survival of the population itself in the face of a likely wide diversity of unpredictable environmental, competitive and hostile challenges ahead. |
|||||||||||||||||||||||||||||||||||||||||||||||||
| Type II TA classification system I: TA family on basis of toxin protein | |||||||||||||||||||||||||||||||||||||||||||||||||
| At present The TA loci in TADB were grouped into
11 two-component and 3 three-component TA families based on toxin protein
sequence similarity. This classification system had been described in the
reviews by Gerdes et al. (Nat Rev Microbiol, 2005, 3(5):371-82) and Van
Melderen et al. (PLoS Genet. 2009, 5(3):e1000437). See Figure S1 in the
supplementary material for a list of all the TA families recorded in TADB.
Through the We have performed pair-wise structure alignments between the ten toxin proteins with known crystal structures using FATCAT (Ye and Godzik, Bioinformatics, 2003, 19(Suppl. 2):ii246-ii255). The FATCAT-defined P-value provides a measure of the chance of two random structures being identified as similar (Table S1). The smaller the P-value, the more statistically significant the similarity between corresponding structures; a pair of proteins with P-value < 5e-02 are considered to be significantly similar. By this criterion, YoeB and MqsR(YgiU) exhibit significant tertiary fold based similarity to RelE. As such the yefM-yoeB family and the ygiTU family were fused with the relBE family. Additionally, as HigB and YhaV have been reported as RelE-homologous (Mol Microbiol. 2010, 75(2):333-48; J Mol Biol. 2007, 372(4):894-905), the higBA and prlF-yhaV families have been fused with the relBE family but as individual subfamilies, given the unique gene order and/or cellular activity associated with each of these relBE subfamilies. Therefore, the relBE TA family now includes five subfamilies: relBE, higBA, yoeB-yefM, ygiTU and prlF-yhaV. However, although ParE also shows significant tertiary fold based similarity to RelE (P-value = 1.29e-06), ParE possesses a unique cellular target and activity (Jiang et al. Mol Microbiol 2002, 44(4):971-9), hence the parDE family has been retained as a separate TA family. Table S1. Tertiary fold-based pair-wise structure alignments of TA system toxin proteins with reported 3-D structures against the RelE toxin (PDB ID: 3KHA.A) using the FATCAT algorithm (a).
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Type II TA classification system II: toxin-antitoxin conserved domain pair | |||||||||||||||||||||||||||||||||||||||||||||||||
| We have also classified TADB entries by a second
toxin-antitoxin domain-based classification system as suggested recently
by Makarova et al. (Biol Direct, 2009, 4:19). Through the Table S2. The relationship of TA family (a) and TA domain pair (b) classification systems of identified and/or predicted TA loci. 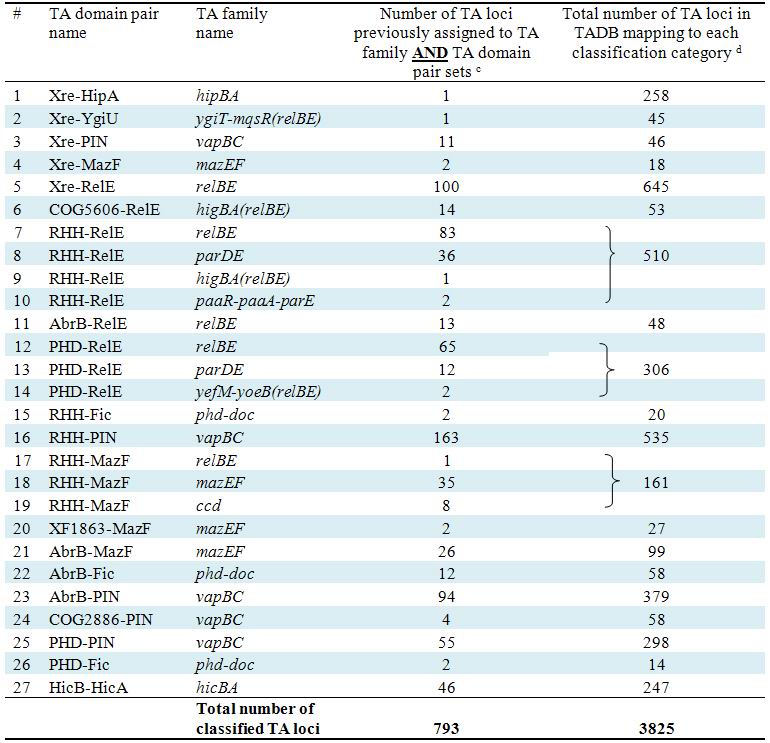
|
|||||||||||||||||||||||||||||||||||||||||||||||||