
| 616 | |
| Enterococcus faecalis plasmid pBEE99 (NC_013533) | |
| plasmid pBEE99 [Browse all T4SS(s) in this replicon] | |
| NC_013533 | |
| 8127..34867 | |
| Prg/Pcf | |
| conjugation | |
| Type IVD; Type GP | |
T4SS components |
| Component | PcfC | PcfH | PrgA | PrgB | PrgC | PrgD | PrgF | PrgH | PrgI |
| Number | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | PrgJ | PrgK | PrgL | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_013533 | ||||||
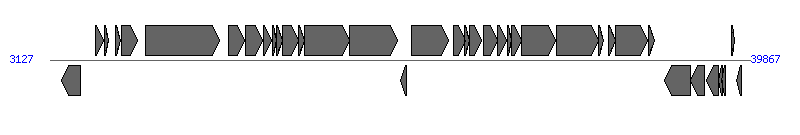 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pBEE99p04 (prgX) | 3731..4714 [-], 984 | 270208276 | PrgX | ||
| 2 | pBEE99p05 (prgR) | 5534..5914 [+], 381 | 270208277 | PrgR | ||
| 3 | pBEE99p06 (prgS) | 5965..6186 [+], 222 | 270208278 | PrgS | ||
| 4 | pBEE99p07 | 6539..6829 [+], 291 | 270208279 | transposase | ||
| 5 | pBEE99p08 | 6865..7701 [+], 837 | 270208280 | integrase | ||
| 6 | pBEE99p09 (prgB) | 8127..12044 [+], 3918 | 270208281 | PrgB | PrgB | |
| 7 | pBEE99p10 (prgC) | 12498..13355 [+], 858 | 270208282 | PrgC | PrgC | |
| 8 | pBEE99p11 (prgD) | 13397..14296 [+], 900 | 270208283 | PrgD | PrgD | |
| 9 | pBEE99p12 (prgK) | 14316..14750 [+], 435 | 270208284 | PrgK | ||
| 10 | pBEE99p13 (prgF) | 14797..15024 [+], 228 | 270208285 | PrgF | PrgF | |
| 11 | pBEE99p14 | 15038..15334 [+], 297 | 270208286 | hypothetical protein | ||
| 12 | pBEE99p15 (prgH) | 15349..16149 [+], 801 | 270208287 | PrgH | PrgH | |
| 13 | pBEE99p16 (prgI) | 16151..16504 [+], 354 | 270208288 | PrgI | PrgI | |
| 14 | pBEE99p17 (prgJ) | 16458..18848 [+], 2391 | 270208289 | PrgJ | PrgJ | |
| 15 | pBEE99p18 (prgK) | 18860..21373 [+], 2514 | 270208290 | PrgK | PrgK | |
| 16 | pBEE99p19 | 21509..21838 [-], 330 | 270208291 | hypothetical protein | ||
| 17 | pBEE99p20 | 22097..24031 [+], 1935 | 270208292 | reverse transcriptase | ||
| 18 | pBEE99p21 (prgL) | 24304..24930 [+], 627 | 270208293 | PrgL | PrgL | |
| 19 | pBEE99p22 (prgM) | 24908..25162 [+], 255 | 270208294 | PrgM | ||
| 20 | pBEE99p23 (pcfA) | 25146..25754 [+], 609 | 270208295 | PcfA | ||
| 21 | pBEE99p24 | 25867..26622 [+], 756 | 270208296 | hypothetical protein | ||
| 22 | pBEE99p25 (pcfS) | 26639..27097 [+], 459 | 270208297 | PcfS | ||
| 23 | pBEE99p26 | 27143..27325 [+], 183 | 270208298 | hypothetical protein | ||
| 24 | pBEE99p27 (pcfB) | 27364..27852 [+], 489 | 270208299 | PcfB | ||
| 25 | pBEE99p28 (pcfC) | 27852..29681 [+], 1830 | 270208300 | PcfC | PcfC | |
| 26 | pBEE99p29 (pcfD) | 29732..31891 [+], 2160 | 270208301 | PcfD | ||
| 27 | pBEE99p30 (pcfE) | 31924..32196 [+], 273 | 270208302 | PcfE | ||
| 28 | pBEE99p31 (pcfF) | 32442..32798 [+], 357 | 270208303 | PcfF | ||
| 29 | pBEE99p32 (pcfG) | 32799..34484 [+], 1686 | 270208304 | PcfG | ||
| 30 | pBEE99p33 (pcfH) | 34517..34867 [+], 351 | 270208305 | PcfH | PcfH | |
| 31 | pBEE99p34 (pcfJ) | 35384..36727 [-], 1344 | 270208306 | PcfJ | ||
| 32 | pBEE99p35 | 36727..37500 [-], 774 | 270208307 | hypothetical protein | ||
| 33 | pBEE99p36 | 37605..38195 [-], 591 | 270208308 | hypothetical protein | ||
| 34 | pBEE99p37 (pcfM) | 38219..38410 [-], 192 | 270208309 | PcfM | ||
| 35 | pBEE99p38 (pcfN) | 38395..38580 [-], 186 | 270208310 | PcfN | ||
| 36 | pBEE99p39 (pcfP) | 38876..39076 [+], 201 | 270208311 | PcfP | ||
| 37 | pBEE99p40 (pcfQ) | 39178..39408 [-], 231 | 270208312 | PcfQ | ||
Download FASTA format files |
| Proteins Genes |
| # | Accessory protein(GI) | motif(s) | Substrate(s) | Function | Reference |
| 1 | PcfF (270208303) | ND (interacts with PcfC T4CP) | PcfG | PcfF interacts with PcfC and PcfG, spatially positions relaxosome or transfer intermediate near the T4CP. | (1) PubMed: 19946141 |
| 2 | PrgB (270208281) | ND | - | Pheromonedependent overproduction of PrgB induces formation of intercellular aggregates. However, upon overproduction, PrgB confers toxicity on E. faecalis donors by a mechanism dependent on extracellular DNA. | (2) PubMed: 25431047 |
| (1) Alvarez-Martinez CE; Christie PJ (2009). Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 73(4):775-808. [PudMed:19946141] |
|
(2) Bhatty M et al. (2015). Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 95(4):660-77. [PudMed:25431047] |
| (1) Coburn PS; Baghdayan AS; Craig N; Burroughs A; Tendolkar P; Miller K; Najar FZ; Roe BA; Shankar N (2010). A novel conjugative plasmid from Enterococcus faecalis E99 enhances resistance to ultraviolet radiation. Plasmid. 64(1):18-25. [PudMed:20307569] |