
| 615 | |
| Enterococcus faecalis plasmid pCF10 (NC_006827) | |
| plasmid pCF10 [Browse all T4SS(s) in this replicon] | |
| NC_006827 | |
| 10015..35356 | |
| Prg/Pcf | |
| conjugation | |
| Type IVD; Type GP | |
T4SS components |
| Component | PcfC | PcfH | PrgA | PrgB | PrgC | PrgD | PrgF | PrgH | PrgI |
| Number | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Component | PrgJ | PrgK | PrgL | ||||||
| Number | 1 | 1 | 1 |
The information of T4SS components from NC_006827 | ||||||
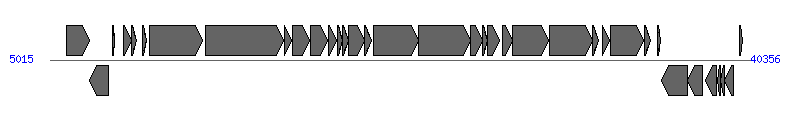 | ||||||
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | pCF10_09 (prgY) | 5843..6997 [+], 1155 | 58616131 | PrgY | ||
| 2 | pCF10_10 (prgX) | 7030..7983 [-], 954 | 58616133 | PrgX | ||
| 3 | pCF10_11 (prgQ) | 8192..8263 [+], 72 | 58616132 | PrgQ | ||
| 4 | pCF10_12 (prgR) | 8745..9125 [+], 381 | 113706805 | PrgR | ||
| 5 | pCF10_13 (prgS) | 9125..9397 [+], 273 | 58616086 | PrgS | ||
| 6 | pCF10_14 (prgT) | 9699..9887 [+], 189 | 58616137 | PrgT | ||
| 7 | pCF10_15 (prgA) | 10015..12690 [+], 2676 | 58616134 | PrgA | PrgA | |
| 8 | pCF10_16 (prgB) | 12883..16800 [+], 3918 | 58616135 | PrgB | PrgB | |
| 9 | pCF10_17 (prgU) | 16862..17218 [+], 357 | 58616087 | PrgU | ||
| 10 | pCF10_18 (prgC) | 17246..18103 [+], 858 | 58616136 | PrgC | PrgC | |
| 11 | pCF10_19 (prgD) | 18145..19044 [+], 900 | 58616088 | PrgD | PrgD | |
| 12 | pCF10_20 (prgE) | 19064..19498 [+], 435 | 58616089 | PrgE | ||
| 13 | pCF10_21 (prgF) | 19545..19772 [+], 228 | 58616090 | PrgF | PrgF | |
| 14 | pCF10_22 (prgG) | 19786..20082 [+], 297 | 58616091 | PrgG | ||
| 15 | pCF10_23 (prgH) | 20097..20897 [+], 801 | 58616084 | PrgH | PrgH | |
| 16 | pCF10_24 (prgI) | 20899..21252 [+], 354 | 58616092 | PrgI | PrgI | |
| 17 | pCF10_25 (prgJ) | 21359..23596 [+], 2238 | 58616093 | PrgJ | PrgJ | |
| 18 | pCF10_26 (prgK) | 23608..26223 [+], 2616 | 58616094 | PrgK | PrgK | |
| 19 | pCF10_27 (prgL) | 26247..26873 [+], 627 | 58616095 | PrgL | PrgL | |
| 20 | pCF10_28 (prgM) | 26860..27105 [+], 246 | 58616096 | PrgM | ||
| 21 | pCF10_29 (pcfA) | 27089..27697 [+], 609 | 58616097 | PcfA | ||
| 22 | pCF10_30 (pcfB) | 27856..28341 [+], 486 | 113706806 | PcfB | ||
| 23 | pCF10_31 (pcfC) | 28341..30170 [+], 1830 | 58616099 | PcfC | PcfC | |
| 24 | pCF10_32 (pcfD) | 30221..32380 [+], 2160 | 58616100 | PcfD | ||
| 25 | pCF10_33 (pcfE) | 32413..32685 [+], 273 | 58616101 | PcfE | ||
| 26 | pCF10_34 (pcfF) | 32931..33287 [+], 357 | 58616102 | PcfF | ||
| 27 | pCF10_35 (pcfG) | 33288..34973 [+], 1686 | 58616103 | PcfG | ||
| 28 | pCF10_36 (pcfH) | 35006..35356 [+], 351 | 58616104 | PcfH | PcfH | |
| 29 | pCF10_37 (pcfI) | 35680..35847 [+], 168 | 58616105 | PcfI | ||
| 30 | pCF10_38 (pcfJ) | 35873..37210 [-], 1338 | 58616106 | PcfJ | ||
| 31 | pCF10_39 (pcfK) | 37207..37983 [-], 777 | 58616107 | PcfK | ||
| 32 | pCF10_40 (pcfL) | 38126..38674 [-], 549 | 58616108 | PcfL | ||
| 33 | pCF10_41 (pcfM) | 38698..38889 [-], 192 | 58616126 | PcfM | ||
| 34 | pCF10_42 (pcfN) | 38874..39059 [-], 186 | 58616109 | PcfN | ||
| 35 | pCF10_43 (pcfO) | 39065..39514 [-], 450 | 58616110 | PcfO | ||
| 36 | pCF10_44 (pcfP) | 39801..40001 [+], 201 | 58616111 | PcfP | ||
Download FASTA format files |
| Proteins Genes |
| # | Name | Image | Resource | Detail | Reference |
| 1 | PcfF | 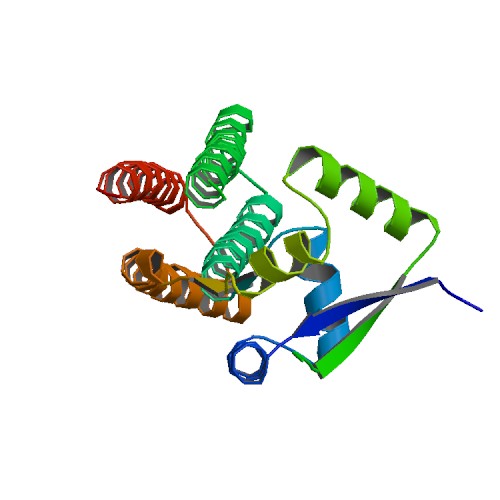 | PDB (6QEQ) | PcfF from Enterococcus faecalis pCF10. | (1) PubMed: 31134011 |
| 2 | PrgB | 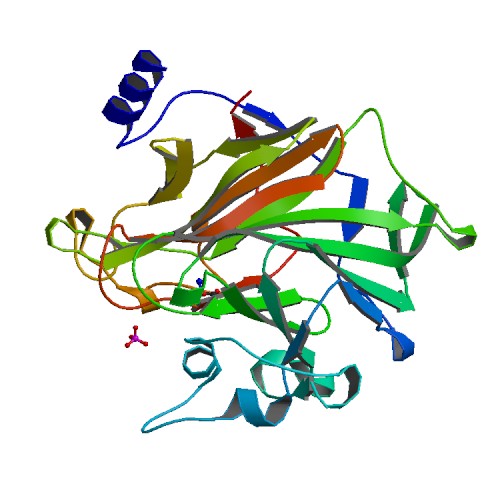 | PDB (6EVU) | Adhesin domain of PrgB from Enterococcus faecalis. | (2) PubMed: 29723434 |
| 3 | PrgB bound to DNA | 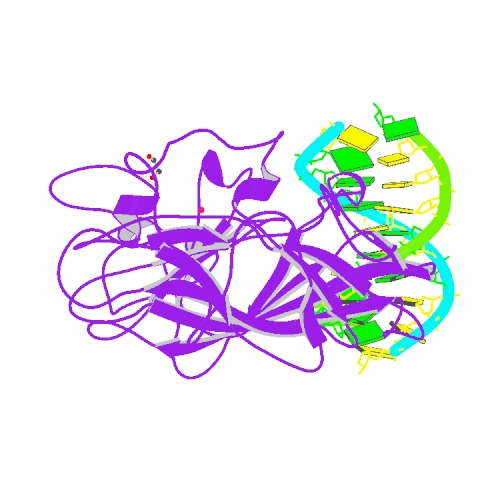 | PDB (6GED) | Adhesin domain of PrgB from Enterococcus faecalis bound to DNA. | (3) PubMed: 29723434 |
|
(1) Rehman S et al. (2019). Enterococcal PcfF Is a Ribbon-Helix-Helix Protein That Recruits the Relaxase PcfG Through Binding and Bending of the oriT Sequence. Front Microbiol. 10:958. [PudMed:31134011] |
|
(2) Schmitt A et al. (2018). PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction. Mol Microbiol. 109(3):291-305. [PudMed:29723434] |
|
(3) Schmitt A et al. (2018). PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction. Mol Microbiol. 109(3):291-305. [PudMed:29723434] |
| # | Accessory protein(GI) | motif(s) | Substrate(s) | Function | Reference |
| 1 | PcfF (58616102) | ND (interacts with PcfC T4CP) | PcfG | PcfF interacts with PcfC and PcfG, spatially positions relaxosome or transfer intermediate near the T4CP. | (1) PubMed: 18811729 |
| 2 | PrgB (58616135) | ND | - | Pheromonedependent overproduction of PrgB induces formation of intercellular aggregates. However, upon overproduction, PrgB confers toxicity on E. faecalis donors by a mechanism dependent on extracellular DNA. | (2) PubMed: 25431047 |
|
(1) Al-Khodor S; Price CT; Habyarimana F; Kalia A; Abu Kwaik Y (2008). A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol. 70(4):908-23. [PudMed:18811729] |
|
(2) Bhatty M et al. (2015). Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol. 95(4):660-77. [PudMed:25431047] |
|
(1) Li F; Alvarez-Martinez C; Chen Y; Choi KJ; Yeo HJ; Christie PJ (2012). Enterococcus faecalis PrgJ, a VirB4-like ATPase, Mediates pCF10 Conjugative Transfer Through Substrate Binding. J Bacteriol. . [PudMed:22636769] |
|
(2) Chen Y; Zhang X; Manias D; Yeo HJ; Dunny GM; Christie PJ (2008). Enterococcus faecalis PcfC, a spatially localized substrate receptor for type IV secretion of the pCF10 transfer intermediate. J Bacteriol. 190(10):3632-45. [PudMed:18326569] |