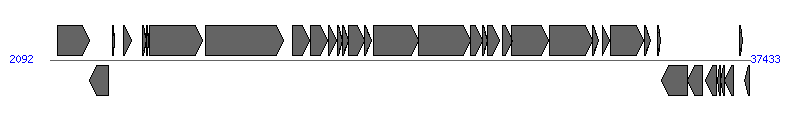The information of T4SS components from NC_004671 |
 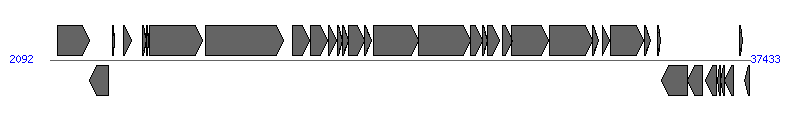 |
| # | Locus tag (Gene) | Coordinates [+/-], size (bp) | Protein GI | Product | Component | |
| 1 | EF_B0004 (traC-2) | 2486..4075 [+], 1590 | 29377898 | TraC protein | | |
| 2 | EF_B0005 | 4101..5042 [-], 942 | 29377899 | Cro/CI family transcriptional regulator | | |
| 3 | EF_B00051 | 5255..5332 [+], 78 | 29377900 | hypothetical protein | | |
| 4 | EF_B0007 | 5822..6202 [+], 381 | 29377901 | pheromone-responsive regulatory protein R | | |
| 5 | EF_B0008 | 6776..6964 [+], 189 | 29377902 | hypothetical protein | | |
| 6 | EF_B0009 | 6968..7081 [+], 114 | 29377903 | hypothetical protein | | |
| 7 | EF_B0010 (prgA) | 7092..9767 [+], 2676 | 29377904 | surface exclusion protein PrgA | PrgA | |
| 8 | EF_B0011 (prgB) | 9960..13877 [+], 3918 | 29377905 | aggregation substance PrgB | PrgB |  |
| 9 | EF_B0012 (prgC) | 14323..15180 [+], 858 | 29377906 | surface protein PrgC | PrgC | |
| 10 | EF_B0013 | 15222..16121 [+], 900 | 29377907 | hypothetical protein | PrgD | |
| 11 | EF_B0014 | 16141..16575 [+], 435 | 29377908 | hypothetical protein | | |
| 12 | EF_B0015 | 16622..16849 [+], 228 | 29377909 | hypothetical protein | PrgF | |
| 13 | EF_B0016 | 16863..17159 [+], 297 | 29377910 | hypothetical protein | | |
| 14 | EF_B0017 | 17174..17974 [+], 801 | 29377911 | hypothetical protein | PrgH | |
| 15 | EF_B0018 | 17976..18329 [+], 354 | 29377912 | hypothetical protein | PrgI | |
| 16 | EF_B0019 | 18436..20673 [+], 2238 | 29377913 | hypothetical protein | PrgJ | |
| 17 | EF_B0020 | 20685..23300 [+], 2616 | 29377914 | hypothetical protein | PrgK | |
| 18 | EF_B0021 | 23324..23950 [+], 627 | 29377915 | hypothetical protein | PrgL | |
| 19 | EF_B0022 | 23928..24182 [+], 255 | 29377916 | hypothetical protein | | |
| 20 | EF_B0023 | 24166..24774 [+], 609 | 29377917 | hypothetical protein | | |
| 21 | EF_B0024 | 24931..25419 [+], 489 | 29377918 | hypothetical protein | | |
| 22 | EF_B0025 | 25416..27248 [+], 1833 | 29377919 | TraG family protein | PcfC | |
| 23 | EF_B0026 | 27298..29457 [+], 2160 | 29377920 | hypothetical protein | | |
| 24 | EF_B0027 | 29490..29762 [+], 273 | 29377921 | LtrD-related protein, putative | | |
| 25 | EF_B0029 | 30008..30364 [+], 357 | 29377922 | hypothetical protein | | |
| 26 | EF_B0030 | 30365..32050 [+], 1686 | 29377923 | relaxase | | |
| 27 | EF_B0031 | 32104..32433 [+], 330 | 29377924 | hypothetical protein | PcfH | |
| 28 | EF_B0032 | 32757..32924 [+], 168 | 29377925 | hypothetical protein | | |
| 29 | EF_B0033 | 32950..34293 [-], 1344 | 29377926 | hypothetical protein | | |
| 30 | EF_B0034 | 34284..35060 [-], 777 | 29377927 | hypothetical protein | | |
| 31 | EF_B0035 | 35203..35751 [-], 549 | 29377928 | hypothetical protein | | |
| 32 | EF_B0036 | 35775..35948 [-], 174 | 29377929 | hypothetical protein | | |
| 33 | EF_B0037 | 35951..36136 [-], 186 | 29377930 | hypothetical protein | | |
| 34 | EF_B0038 | 36142..36591 [-], 450 | 29377931 | hypothetical protein | | |
| 35 | EF_B0040 | 36878..37078 [+], 201 | 29377932 | hypothetical protein | | |
| 36 | EF_B0041 | 37179..37409 [-], 231 | 29377933 | hypothetical protein | | |
| |