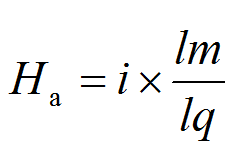The Tutorial on the KpVR Tool
A Web tool for detecting replicons, virulence, T4SS gene clusters and antibiotic resistance genes in K. pneumoniae plasmids. KpVR was designed to show the correlation between replicons with virulence and conjugative modules, for tracking the evolution of virulence plasmids. We first developed a back-end dataset repDB based on our IncFIBK subtypes. The online tool KpVR then performs rapid homology searches of a query plasmid sequence against repDB based on nucleotide sequence identities and coverage. Furthermore, iuc and iro lineages based on the AbST and SmST typing schemes were also integrated by KpVR. KpVR outputs a simple list and generates a graphic overview of not only the prediction of replicon types but also the extended putative virulence modules or acquired antibiotic resistance genes.
| 1. How to use KpVR |
| 1.1. Input a Nucleotide Sequence |
In the first place, you should specify the sequence which you want to analysis. Single or multiple sequence in FASTA Format is acceptable to this server, and a warning will be given when sequence in wrong format be found, the sequence ids should not contain any special characters.
If you have trouble in choosing sequence, the web page also provides an example. Please click the "Show an example" to provide an example for KpVR.
|
| 1.2. Choose the prediction Parameters |
For this option, there are three optional parameters, which are provided for users to choose for prediction of replicons, virulence, T4SS gene cluster and antibiotic resistance genes based on input sequences. |
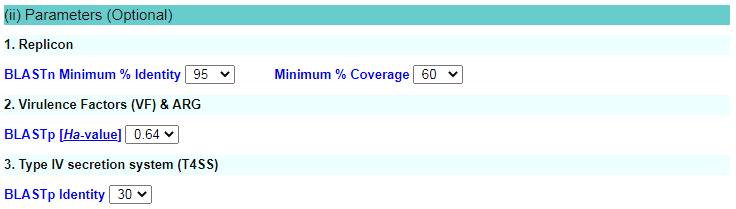 |
| Use the Select threshold for Minimum % Identities check box to select the minimum percentage of nucleotides that are identical between the best matching sequence in the repDB and the corresponding sequence in the plasmid genome. If "Identities threshold" is equal to or larger than the selected threshold will be shown in the output. By default the Minimum % Identities = 95. |
Use the Select Minimum % Coverage check box to select the minimum percentage of coverage between uploaded sequence and repDB. If "Coverage threshold" is equal to or larger than the selected threshhold will be shown in the output. By default the Minimum % Coverage = 60. |
To examine the degree of sequence similarities at an amino acid level between query protein and the dataset of virulence and resistance genes, the NCBI BLASTp-derived Ha-value was employed. For each query, the Ha-value was calculated as follows:
Among them, i is the BLASTp identity level of the region with the highest bit score, representing the frequency between 0 and 1, lm is the length of the matching sequence (including the gaps) with the highest score, and lq is the query length. If there is no matching sequence with BLASTp E-value < 0.001, the Ha-value assigned to the query sequence is defined as zero. So Ha-value belongs to set, Ha ∈[0,1]. Here, the strict Ha-value cut-off 0.64 is used to determine the significant sequence similarity; for example, the identities is 80% and the ratio of matching length is 80%.
|
| Use the Select threshold for identities drop-down menu to select the Minimum identities of proteins that are identical between the best matching genes in the T4SS dataset and the query genes in the plasmid. All genes with identities value equal or larger than the selected threshold will be shown in the output. By default the Minimum %identities = 30. |
| 2. Interpreting the prediction output |
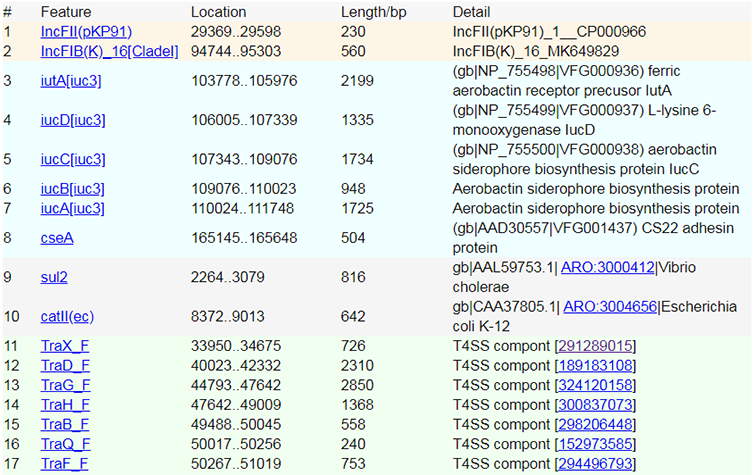
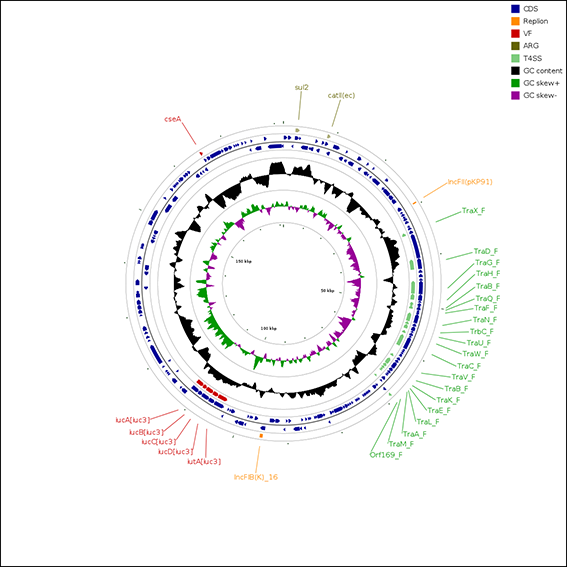
When a prediction task has been finished, you can obtain your query results. The following is a screenshot of a result page.
The prediction of different feature results are shown in a table and a graphic overview with differnet colors. |