The graph information of IME_SagA909_tRNAlys components from CP000114 |
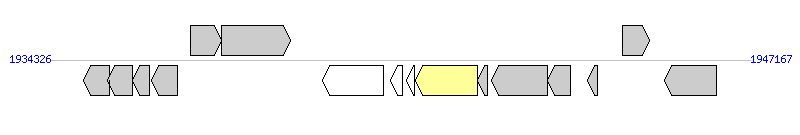 |
| Incomplete gene list of IME_SagA909_tRNAlys from CP000114 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | SAK_1933 | 1934944..1935414 [-], 471 | acetyltransferase, GNAT family | |
| 2 | SAK_1934 | 1935386..1935844 [-], 459 | hydrolase, NUDIX family | |
| 3 | SAK_1935 | 1935837..1936145 [-], 309 | conserved hypothetical protein | |
| 4 | SAK_1936 | 1936188..1936658 [-], 471 | conserved hypothetical protein | |
| 5 | SAK_1937 | 1936894..1937475 [+], 582 | acetyltransferase, GNAT family | |
| 6 | SAK_1938 | 1937468..1938736 [+], 1269 | ATPase, AAA family | |
| 7 | SAK_1940 | 1939326..1940435 [-], 1110 | hypothetical protein | |
| 8 | SAK_1941 | 1940575..1940784 [-], 210 | hypothetical protein | |
| 9 | SAK_1942 | 1940873..1941013 [-], 141 | hypothetical protein | |
| 10 | SAK_1943 | 1941036..1942166 [-], 1131 | site-specific recombinase, phage integrase family | Integrase |
| 11 | SAK_1944 | 1942170..1942355 [-], 186 | conserved hypothetical protein | |
| 12 | SAK_1945 | 1942416..1943444 [-], 1029 | replication initiation factor family protein | |
| 13 | SAK_1946 | 1943447..1943866 [-], 420 | conserved domain protein | |
| 14 | SAK_1947 | 1944192..1944365 [-], 174 | hypothetical protein | |
| 15 | SAK_1948 | 1944822..1945316 [+], 495 | transcriptional regulator, putative | |
| 16 | SAK_1949 | 1945603..1946553 [-], 951 | conserved hypothetical protein | |
