The graph information of Tn6104 components from FN545816 |
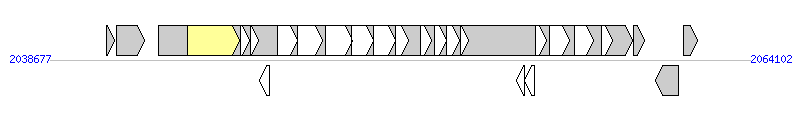 |
| Complete gene list of Tn6104 from FN545816 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | CDR20291_1741 | 2040727..2041008 [+], 282 | putative membrane protein precursor | |
| 2 | CDR20291_1742 | 2041094..2042116 [+], 1023 | putative conjugative transposon protein | |
| 3 | CDR20291_1743 | 2042600..2059823 [+], 17224 | putative conjugal transfer protein pseudogene | |
| 4 | CDR20291_1744 | 2043677..2045557 [+], 1881 | site-specific recombinase, resolvase family, putative | Integrase |
| 5 | CDR20291_1745 | 2045584..2045937 [+], 354 | putative uncharacterized protein | |
| 6 | CDR20291_1746 | 2045945..2046253 [+], 309 | putative uncharacterized protein | |
| 7 | CDR20291_1747 | 2046281..2046649 [-], 369 | putative conjugative transposon regulatory protein | |
| 8 | CDR20291_1748 | 2046924..2047667 [+], 744 | two-component response regulator | |
| 9 | CDR20291_1749 | 2047673..2048590 [+], 918 | sensor protein | |
| 10 | CDR20291_1750 | 2048689..2049651 [+], 963 | putative lantibiotic abc transporter,atp-binding protein | |
| 11 | CDR20291_1751 | 2049644..2050429 [+], 786 | putative lantibiotic abc transporter, permease protein | |
| 12 | CDR20291_1752 | 2050432..2051211 [+], 780 | putative lantibiotic abc transporter, permease protein | |
| 13 | CDR20291_1753 | 2051219..2051680 [+], 462 | putative uncharacterized protein | |
| 14 | CDR20291_1754 | 2052125..2052538 [+], 414 | rna polymerase, sigma-24 subunit, ecf subfamily (ecf subfamily rna polymerase sigma-70 factor) | |
| 15 | CDR20291_1755 | 2052640..2053092 [+], 453 | sigma-24 (feci) | |
| 16 | CDR20291_1756 | 2053067..2053564 [+], 498 | rna polymerase, sigma-24 subunit, ecf subfamily | |
| 17 | CDR20291_1757 | 2053584..2053883 [+], 300 | putative uncharacterized protein | |
| 18 | CDR20291_1759 | 2055623..2055913 [-], 291 | addiction module toxin, rele/stbe family | |
| 19 | CDR20291_1760 | 2055910..2056278 [-], 369 | addiction module antitoxin, relb/dinj family | |
| 20 | CDR20291_1761 | 2056315..2056713 [+], 399 | putative uncharacterized protein | |
| 21 | CDR20291_1762 | 2056827..2057561 [+], 735 | phage protein | |
| 22 | CDR20291_1763 | 2057726..2058421 [+], 696 | replicative dna helicase | |
| 23 | CDR20291_1764 | 2058698..2059102 [+], 405 | putative uncharacterized protein | |
| 24 | CDR20291_1743 | 2042600..2059823 [+], 17224 | putative conjugal transfer protein pseudogene | |
| 25 | CDR20291_1765 | 2059876..2060253 [+], 378 | putative uncharacterized protein | |
| 26 | yobD | 2060670..2061518 [-], 849 | transcription regulator (yobd protein) | |
| 27 | CDR20291_1767 | 2061695..2062177 [+], 483 | putative uncharacterized protein | |
