
| 239_IME | |
| Tn4399 | |
| - | |
| Bacteroides fragilis TM4.2321 | |
| 9.6 kb | |
| Numerous sites | |
| - | |
| - | |
| L20975.1 (partical IME sequence) | |
| coordinates: 991..1189; oriTDB id: 200009 GATTCGGCATCTAAGCCGTAAATCACATCTTTTTTAGGATAGCAAGTTAATGTCGGCGTATTACATTCCC CGACCACTTGCCCCTCAAAAAGATGAGGGGAACGGCTCCCGATGGTCACGGACTTTTCAGATTACATCAA TCGGATTATTCACTAAAAAACAGATAGAACAGATGAAGAAAAACAACATAATCACCTTA | |
| coordinates: 1461..2417; Locus tag: -; Family: MOBP |
The Interaction Network among ICE/IME/CIME/plasmid | ||||||
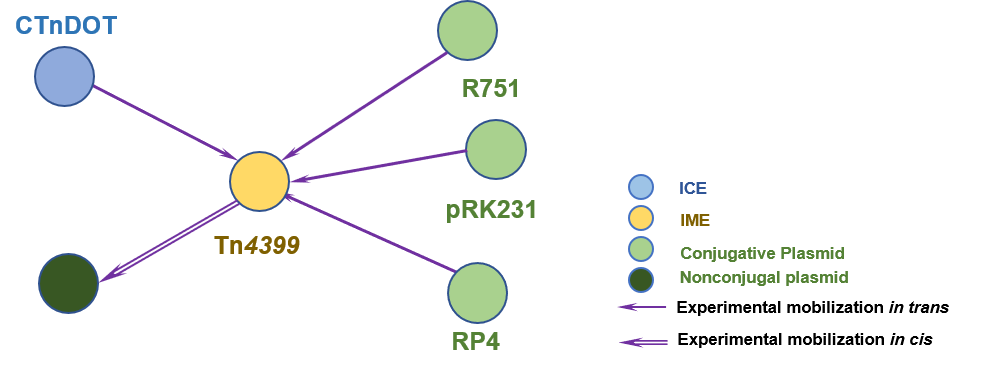 | ||||||
| Detailed Informatioin of the Interaction Network | ||||||
| # | IME | Inter_Ele | Methods | Donors | Recipients | Exper_Ref |
| 1 | Tn4399 | CTnDOT [ICE] | | Bacteroides | Bacteroides | 8397185 |
| 2 | Tn4399 | pRK231 [IncP plasmid] | | Escherichia coli | Escherichia coli | 8397185 |
| 3 | Tn4399 | RP4 [IncP plasmid] | | Escherichia coli | Escherichia coli | 8397185 |
| 4 | Tn4399 | R751 [IncP plasmid] | | Escherichia coli | Escherichia coli | 8397185 |
| 5 | Tn4399 | pGAT400ΔBglII [nonconjugal plasmid] | | Bacteroides fragilis | Escherichia coli; Bacteroides fragilis | 2544548 |