The graph information of SGI1-V components from HQ888851 |
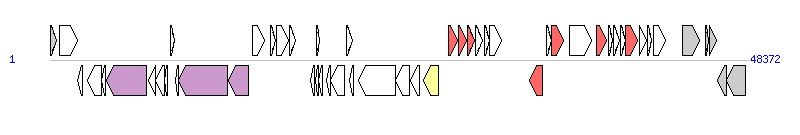 |
| Complete gene list of SGI1-V from HQ888851 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | thdF | 1..468 [+], 468 | tRNA modification GTPase | |
| 2 | int | 672..1889 [+], 1218 | integrase | |
| 3 | xis | 1886..2254 [-], 369 | excisionase | |
| 4 | rep | 2612..3565 [-], 954 | replication protein | |
| 5 | - | 3552..3842 [-], 291 | hypothetical protein | |
| 6 | - | 3930..6689 [-], 2760 | conjugal transfer mating pair stabilization protein TraN | TraN_F, T4SS component |
| 7 | - | 6786..7319 [-], 534 | putative regulator protein | |
| 8 | - | 7322..7933 [-], 612 | hypothetical protein | |
| 9 | - | 7933..8154 [-], 222 | hypothetical protein | |
| 10 | - | 8329..8619 [+], 291 | hypothetical protein | |
| 11 | - | 8639..8893 [-], 255 | hypothetical protein | |
| 12 | - | 8893..12297 [-], 3405 | conjugative transfer pilus assembly protein TraG | TraG_F, T4SS component |
| 13 | - | 12301..13725 [-], 1425 | conjugal transfer pilus assembly protein TraH | TraH_F, T4SS component |
| 14 | - | 13969..14826 [+], 858 | hypothetical protein | |
| 15 | - | 15257..15571 [+], 315 | hypothetical protein | |
| 16 | - | 15681..16472 [+], 792 | hydrolase-like protein | |
| 17 | - | 16565..16984 [+], 420 | hypothetical protein | |
| 18 | - | 17990..18259 [-], 270 | hypothetical protein | |
| 19 | - | 18337..18522 [-], 186 | hypothetical protein | |
| 20 | - | 18382..18636 [+], 255 | hypothetical protein | |
| 21 | - | 18533..18850 [-], 318 | hypothetical protein | |
| 22 | - | 19109..19405 [-], 297 | hypothetical protein | |
| 23 | - | 19409..20374 [-], 966 | phage integrase family protein | |
| 24 | - | 20467..20928 [+], 462 | S021-like elongation protein | |
| 25 | - | 20714..20989 [-], 276 | hypothetical protein | |
| 26 | - | 21322..23865 [-], 2544 | subtilisin-like peptidase | |
| 27 | - | 23886..24875 [-], 990 | AAA ATPase, central domain protein | |
| 28 | res | 24945..25529 [-], 585 | resolvase | |
| 29 | intI1 | 25802..26815 [-], 1014 | IntI1 integrase | Integrase |
| 30 | accA4 | 27564..28169 [+], 606 | AacA4 aminoglycoside (6') acetyltransferase | AR |
| 31 | aadB | 28239..28772 [+], 534 | AadB aminoglycoside (2'') adenylyltransferase | AR |
| 32 | dhfrA1 | 28841..29314 [+], 474 | DhfrA1 dihydrofolate reductase | AR |
| 33 | - | 29408..29887 [+], 480 | hypothetical protein | |
| 34 | qacEdelta1 | 30018..30365 [+], 348 | QacEdelta1 | |
| 35 | sul1 | 30359..31198 [+], 840 | Sul1 dihydropteroate synthase | |
| 36 | blaVEB-6 | 33148..34047 [-], 900 | VEB-6 extended-spectrum beta-lactamase | AR |
| 37 | qacEdelta1 | 34296..34643 [+], 348 | QacEdelta1 | |
| 38 | sul1 | 34637..35476 [+], 840 | dihydropteroate synthase | AR |
| 39 | - | 35881..37422 [+], 1542 | transposase | |
| 40 | qnrA1 | 37773..38429 [+], 657 | QnrA1 pentapeptide repeat protein | AR |
| 41 | - | 38573..38872 [+], 300 | hypothetical protein | |
| 42 | - | 38916..39350 [+], 435 | hypothetical protein | |
| 43 | qacEdelta1 | 39444..39791 [+], 348 | QacEdelta1 | |
| 44 | sul1 | 39785..40624 [+], 840 | dihydropteroate synthase | AR |
| 45 | - | 40752..41252 [+], 501 | acetyltransferase | |
| 46 | - | 41276..41563 [+], 288 | hypothetical protein | |
| 47 | tnpA6100 | 41729..42523 [+], 795 | IS6100 transposase | |
| 48 | - | 43692..44909 [+], 1218 | putative phage integrase | |
| 49 | - | 45303..45521 [+], 219 | putative transcriptional regulator | |
| 50 | - | 45562..46089 [+], 528 | hypothetical protein | |
| 51 | - | 46093..46776 [-], 684 | hypothetical protein | |
| 52 | - | 46780..48033 [-], 1254 | hypothetical protein | |