The graph information of SGI1-K7 components from MF372717 |
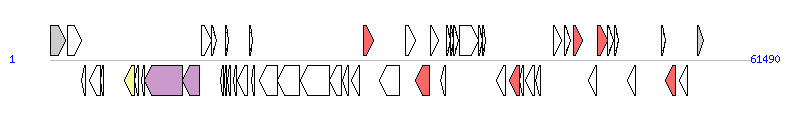 |
| Complete gene list of SGI1-K7 from MF372717 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | - | 1..1365 [+], 1365 | tRNA modification GTPase | |
| 2 | int SGI | 1569..2786 [+], 1218 | integrase | |
| 3 | xis | 2783..3151 [-], 369 | excisionase | |
| 4 | rep | 3509..4462 [-], 954 | replication protein | |
| 5 | - | 4449..4739 [-], 291 | hypothetical protein | |
| 6 | - | 6551..7423 [-], 873 | IS1359 transposase | Integrase |
| 7 | - | 7420..7764 [-], 345 | IS1359 transposase | |
| 8 | - | 8015..8269 [-], 255 | hypothetical protein | |
| 9 | traG | 8269..11673 [-], 3405 | putative pilus assembly protein TraG | TraG_F, T4SS component |
| 10 | traH | 11677..13101 [-], 1425 | conjugal transfer pilus assembly protein TraH | TraH_F, T4SS component |
| 11 | - | 13345..14202 [+], 858 | hypothetical protein | |
| 12 | - | 14199..14594 [+], 396 | hypothetical protein | |
| 13 | - | 15009..15278 [-], 270 | hypothetical protein | |
| 14 | - | 15356..15541 [-], 186 | hypothetical protein | |
| 15 | - | 15401..15655 [+], 255 | hypothetical protein | |
| 16 | - | 15552..15869 [-], 318 | hypothetical protein | |
| 17 | - | 16127..16423 [-], 297 | hypothetical protein | |
| 18 | - | 16427..17392 [-], 966 | putative integrase | |
| 19 | - | 17485..17820 [+], 336 | hypothetical protein | |
| 20 | - | 17732..18007 [-], 276 | hypothetical protein | |
| 21 | - | 18368..20008 [-], 1641 | putative helicase | |
| 22 | - | 20026..21951 [-], 1926 | putative exonuclease | |
| 23 | sgiT | 22029..24572 [-], 2544 | toxin protein of the Toxin-Antitoxin system sgiAT, subtilisin-like serine protease | |
| 24 | sgiA | 24593..25582 [-], 990 | antitoxin protein of the Toxin-Antitoxin system sgiAT, AAA ATPase | |
| 25 | resG | 25661..26245 [-], 585 | resolvase | |
| 26 | tnp26 | 26455..27159 [-], 705 | Tnp26 | |
| 27 | strB | 27566..28402 [+], 837 | streptomycin phosphotransferase B | AR |
| 28 | tnpA' | 28941..30737 [-], 1797 | transposase | |
| 29 | pecM | 31226..32110 [+], 885 | PecM transporter protein | |
| 30 | tetA(A) | 32142..33341 [-], 1200 | TetA(A) | AR |
| 31 | tetR(A) | 33447..34097 [+], 651 | TetR(A) | |
| 32 | merR | 34303..34737 [-], 435 | MerR | |
| 33 | merT | 34809..35159 [+], 351 | MerT | |
| 34 | merP | 35173..35448 [+], 276 | MerP | |
| 35 | merC | 35484..35906 [+], 423 | MerC | |
| 36 | merA | 35958..37640 [+], 1683 | MerA | |
| 37 | merD | 37658..38023 [+], 366 | MerD | |
| 38 | merE | 38020..38256 [+], 237 | MerE | |
| 39 | tnpA6100 | 39262..40056 [-], 795 | IS6100 transposase | |
| 40 | sul1 | 40400..41239 [-], 840 | Sul1 dihydropteroate synthase | AR |
| 41 | qacEdelta1 | 41233..41580 [-], 348 | QacEdelta1 | |
| 42 | aadA7 | 41744..42541 [-], 798 | aminoglycoside (3'')(9) adenylyltransferase | |
| 43 | aacCA5 | 42617..43093 [-], 477 | aminoglycoside (3) acetyltransferase | |
| 44 | tnp26 | 44263..44967 [+], 705 | Tnp26 | |
| 45 | tnpR | 45206..45763 [+], 558 | Tn2 resolvase | |
| 46 | blaTEM1b | 45945..46805 [+], 861 | BlaTEM-1b beta lactamase | AR |
| 47 | tnp26 | 47266..47970 [-], 705 | Tnp26 | |
| 48 | aac(3)-IIa | 48077..48937 [+], 861 | Aminoglycoside 3-N-acetyltransferase, AacC2 | AR |
| 49 | tmrB | 48950..49492 [+], 543 | tunicamycin resistance protein | |
| 50 | - | 49563..49895 [+], 333 | transposase orfA of ISKpn11-like, IS3 family | |
| 51 | tnp26 | 50686..51390 [-], 705 | Tnp26 | |
| 52 | - | 53712..54044 [+], 333 | Tryptophan synthase subunit beta like protein | |
| 53 | blaCTX-M-15 | 54091..54966 [-], 876 | BlaCTX-M-15 beta-lactamase | AR |
| 54 | tnp26 | 55276..55980 [-], 705 | Tnp26 | |
| 55 | - | 56917..57381 [+], 465 | membrane protein | |
