The graph information of SGI1-I components from KJ186152 |
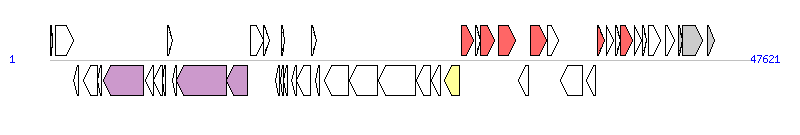 |
| Complete gene list of SGI1-I from KJ186152 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | thdF | 1..162 [+], 162 | tRNA modification GTPase | |
| 2 | int | 366..1583 [+], 1218 | integrase | |
| 3 | xis | 1580..1948 [-], 369 | excisionase | |
| 4 | rep | 2306..3259 [-], 954 | replication protein | |
| 5 | - | 3246..3536 [-], 291 | hypothetical protein | |
| 6 | - | 3624..6383 [-], 2760 | putative mating pair stabilization protein | TraN_F, T4SS component |
| 7 | - | 6480..7013 [-], 534 | putative regulator protein | |
| 8 | - | 7016..7627 [-], 612 | hypothetical protein | |
| 9 | - | 7627..7848 [-], 222 | hypothetical protein | |
| 10 | - | 8023..8313 [+], 291 | hypothetical protein | |
| 11 | - | 8333..8587 [-], 255 | hypothetical protein | |
| 12 | - | 8587..11991 [-], 3405 | putative pilus assembly protein | TraG_F, T4SS component |
| 13 | - | 11995..13419 [-], 1425 | putative pilus assembly protein | TraH_F, T4SS component |
| 14 | - | 13663..14520 [+], 858 | hypothetical protein | |
| 15 | - | 14517..14912 [+], 396 | hypothetical protein | |
| 16 | - | 15327..15596 [-], 270 | hypothetical protein | |
| 17 | - | 15674..15859 [-], 186 | hypothetical protein | |
| 18 | - | 15719..15973 [+], 255 | hypothetical protein | |
| 19 | - | 15870..16187 [-], 318 | hypothetical protein | |
| 20 | - | 16445..16741 [-], 297 | hypothetical protein | |
| 21 | - | 16745..17710 [-], 966 | putative integrase | |
| 22 | - | 17803..18138 [+], 336 | hypothetical protein | |
| 23 | - | 18050..18325 [-], 276 | hypothetical protein | |
| 24 | - | 18686..20326 [-], 1641 | putative helicase | |
| 25 | - | 20344..22269 [-], 1926 | putative exonuclease | |
| 26 | - | 22347..24890 [-], 2544 | putative subtilisin proteinase-like protein | |
| 27 | - | 24911..25900 [-], 990 | putative ATPase | |
| 28 | res | 25979..26563 [-], 585 | resolvase | |
| 29 | intI1 | 26837..27850 [-], 1014 | integrase | Integrase |
| 30 | aadA2 | 27996..28787 [+], 792 | streptomycin/spectinomycin resistance protein | AR |
| 31 | qacEdelta1 | 28951..29298 [+], 348 | quaternary ammonium compound and disinfectant protein | |
| 32 | - | 29292..30263 [+], 972 | sul1delta fusion protein | AR |
| 33 | flo | 30480..31694 [+], 1215 | chloramphenicol and florfenicol resistance protein | AR |
| 34 | tetR | 31901..32527 [-], 627 | tetracycline resistance regulator protein | |
| 35 | tet(G) | 32679..33806 [+], 1128 | tetracycline resistance protein | AR |
| 36 | - | 33827..34618 [+], 792 | putative LysR-type transcriptional regulator | |
| 37 | - | 34710..36194 [-], 1485 | putative transposase-like protein | |
| 38 | groEL/intI1 | 36469..37122 [-], 654 | GroEL/integrase fusion protein | |
| 39 | dfrA1 | 37279..37752 [+], 474 | dihydrofolate reductase I | AR |
| 40 | - | 37846..38325 [+], 480 | OrfC | |
| 41 | qacEdelta1 | 38456..38803 [+], 348 | quaternary ammonium compound and disinfectant protein | |
| 42 | sul1 | 38797..39636 [+], 840 | sulfonamide resistance protein | AR |
| 43 | - | 39764..40264 [+], 501 | putative acetyltransferase | |
| 44 | - | 40288..40575 [+], 288 | hypothetical protein | |
| 45 | tnpA | 40741..41535 [+], 795 | transposase | |
| 46 | - | 41883..42512 [+], 630 | hypothetical protein | |
| 47 | hipB | 42788..43045 [+], 258 | transcriptional regulator | |
| 48 | hipA | 43046..44359 [+], 1314 | regulatory protein | |
| 49 | - | 44762..45204 [+], 443 | membrane protein | |