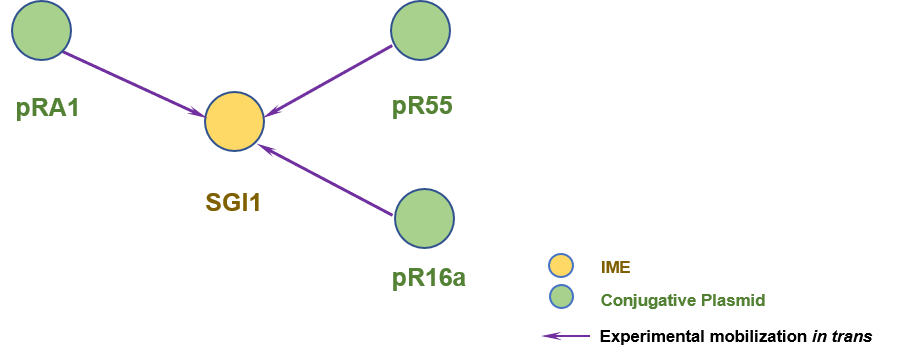
The graph information of SGI1 components from AF261825 |
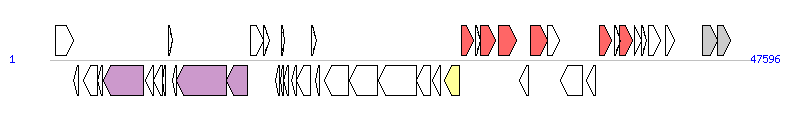 |
| Complete gene list of SGI1 from AF261825 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | int | 367..1584 [+], 1218 | integrase | |
| 2 | xis | 1581..1949 [-], 369 | excisionase | |
| 3 | rep | 2307..3260 [-], 954 | replication protein | |
| 4 | S004 | 3247..3537 [-], 291 | hypothetical protein | |
| 5 | S005 | 3625..6384 [-], 2760 | putative mating pair stabilization protein | TraN_F, T4SS component |
| 6 | S006 | 6481..7014 [-], 534 | putative regulator protein | |
| 7 | S007 | 7017..7628 [-], 612 | hypothetical protein | |
| 8 | S008 | 7628..7849 [-], 222 | hypothetical protein | |
| 9 | S009 | 8024..8314 [+], 291 | hypothetical protein | |
| 10 | - | 8334..8588 [-], 255 | hypothetical protein | |
| 11 | S011 | 8588..11992 [-], 3405 | putative pilus assembly protein | TraG_F, T4SS component |
| 12 | S012 | 11996..13420 [-], 1425 | putative pilus assembly protein | TraH_F, T4SS component |
| 13 | S013 | 13664..14521 [+], 858 | hypothetical protein | |
| 14 | S014 | 14518..14913 [+], 396 | hypothetical protein | |
| 15 | S015 | 15328..15597 [-], 270 | hypothetical protein | |
| 16 | S016 | 15675..15860 [-], 186 | hypothetical protein | |
| 17 | S017 | 15720..15974 [+], 255 | hypothetical protein | |
| 18 | S018 | 15871..16188 [-], 318 | hypothetical protein | |
| 19 | S019 | 16447..16743 [-], 297 | hypothetical protein | |
| 20 | S020 | 16747..17712 [-], 966 | putative integrase | |
| 21 | S021 | 17805..18140 [+], 336 | hypothetical protein | |
| 22 | S022 | 18052..18327 [-], 276 | hypothetical protein | |
| 23 | S023 | 18688..20328 [-], 1641 | putative helicase | |
| 24 | S024 | 20346..22271 [-], 1926 | putative exonuclease | |
| 25 | S025 | 22349..24892 [-], 2544 | putative subtilisin proteinase-like protein | |
| 26 | S026 | 24913..25902 [-], 990 | putative ATPase | |
| 27 | res | 25981..26565 [-], 585 | resolvase | |
| 28 | intI1 | 26839..27852 [-], 1014 | integrase | Integrase |
| 29 | aadA2 | 27998..28789 [+], 792 | streptomycin/spectinomycin resistance protein | AR |
| 30 | qacEdelta1 | 28953..29300 [+], 348 | quaternary ammonium compound and disinfectant protein | |
| 31 | - | 29294..30265 [+], 972 | sul1delta fusion protein | AR |
| 32 | flo | 30482..31696 [+], 1215 | chloramphenicol and florfenicol resistance protein | AR |
| 33 | tetR | 31903..32529 [-], 627 | tetracycline resistance regulator protein | |
| 34 | tet(G) | 32681..33808 [+], 1128 | tetracycline resistance protein | AR |
| 35 | qacEdelta1 | 33829..34620 [+], 792 | putative LysR-type transcriptional regulator | |
| 36 | - | 34712..36196 [-], 1485 | putative transposase-like protein | |
| 37 | groEL/intI1 | 36471..37124 [-], 654 | GroEL/integrase fusion protein | |
| 38 | pse-1 | 37330..38196 [+], 867 | beta-lactamase | AR |
| 39 | qacEdelta1 | 38413..38760 [+], 348 | quaternary ammonium compound and disinfectant protein | |
| 40 | sul1 | 38754..39593 [+], 840 | sulfonamide resistance protein | AR |
| 41 | - | 39721..40221 [+], 501 | putative acetyltransferase | |
| 42 | - | 40245..40532 [+], 288 | hypothetical protein | |
| 43 | tnpA | 40698..41492 [+], 795 | transposase | |
| 44 | S044 | 41840..42469 [+], 630 | hypothetical protein | |
| 45 | urt | 44397..45359 [+], 963 | hypothetical protein | |
| 46 | rt | 45359..46294 [+], 936 | reverse transcriptase | |