The graph information of PGI2 components from MG201402 |
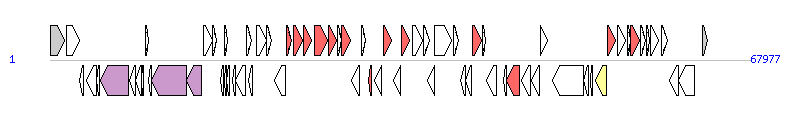 |
| Complete gene list of PGI2 from MG201402 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | trmE | 53..1417 [+], 1365 | tRNA modification GTPase | |
| 2 | int | 1620..2843 [+], 1224 | PGI2 integrase | |
| 3 | xis | 2840..3208 [-], 369 | PGI2 excisionase | |
| 4 | rep | 3563..4516 [-], 954 | PGI2 Rep | |
| 5 | PGI2-004 | 4503..4793 [-], 291 | hypothetical protein | |
| 6 | PGI2-005 | 4881..7640 [-], 2760 | hypothetical protein | TraN_F, T4SS component |
| 7 | PGI2-006 | 7737..8270 [-], 534 | hypothetical protein | |
| 8 | PGI2-007 | 8273..8884 [-], 612 | hypothetical protein | |
| 9 | PGI2-008 | 8884..9105 [-], 222 | hypothetical protein | |
| 10 | PGI2-009 | 9280..9570 [+], 291 | hypothetical protein | |
| 11 | PGI2-010 | 9590..9844 [-], 255 | hypothetical protein | |
| 12 | traG | 9844..13248 [-], 3405 | TraG | TraG_F, T4SS component |
| 13 | traH | 13252..14676 [-], 1425 | TraH | TraH_F, T4SS component |
| 14 | PGI2-013 | 14920..15777 [+], 858 | hypothetical protein | |
| 15 | PGI2-014 | 15774..16169 [+], 396 | hypothetical protein | |
| 16 | PGI2-015 | 16584..16853 [-], 270 | hypothetical protein | |
| 17 | PGI2-016 | 16931..17116 [-], 186 | hypothetical protein | |
| 18 | PGI2-017 | 16976..17230 [+], 255 | hypothetical protein | |
| 19 | PGI2-018 | 17127..17444 [-], 318 | hypothetical protein | |
| 20 | PGI2-019 | 17702..17998 [-], 297 | hypothetical protein | |
| 21 | PGI2-020 | 18002..18967 [-], 966 | Int | |
| 22 | PGI2-021 | 19060..19521 [+], 462 | hypothetical protein | |
| 23 | PGI2-022 | 19307..19627 [-], 321 | hypothetical protein | |
| 24 | PGI2-023 | 20071..21006 [+], 936 | hypothetical protein | |
| 25 | PGI2-024 | 21003..21539 [+], 537 | hypothetical protein | |
| 26 | intI1 | 21812..22825 [-], 1014 | IntI1 | |
| 27 | dfrA16 | 22981..23454 [+], 474 | dihydrofolate reductase | AR |
| 28 | blaPSE-1 | 23619..24485 [+], 867 | PSE-1 beta-lactamase | AR |
| 29 | aadA2 | 24603..25394 [+], 792 | streptomycin/spectinomycin 3' adenyltransferase | AR |
| 30 | cmlA1 | 25656..26915 [+], 1260 | chloramphenicol efflux pump | AR |
| 31 | aadA1 | 27008..27799 [+], 792 | aminoglycoside-3'-adenylyltransferase | AR |
| 32 | qacEdelta1_1 | 27963..28310 [+], 348 | quaternary ammonium compound resistance protein | |
| 33 | sul1_1 | 28304..29143 [+], 840 | Sul1 | AR |
| 34 | IS26 tnpA_1 | 29307..30011 [-], 705 | IS26 transposase | |
| 35 | PGI2-034 | 30207..30599 [+], 393 | NimC/NimA family protein | |
| 36 | blms | 30919..31203 [-], 285 | Bleomycin resistance protein | AR |
| 37 | IS26 tnpA_2 | 31499..32203 [-], 705 | IS26 transposase | |
| 38 | aphA1 | 32354..33169 [+], 816 | aminoglycoside phosphotransferase | AR |
| 39 | IS26 tnpA_3 | 33359..34063 [-], 705 | IS26 transposase | |
| 40 | aac(3)-IV | 34150..34953 [+], 804 | aminoglycoside acetyltransferase | AR |
| 41 | hph | 35182..36207 [+], 1026 | hygromycin B phosphotransferase Hph | |
| 42 | PGI2-041 | 36282..36863 [+], 582 | hypothetical protein | |
| 43 | PGI2-042 | 36629..37381 [-], 753 | hypothetical protein | |
| 44 | Tn3 tnpA | 37365..38966 [+], 1602 | transposase of Tn3 | |
| 45 | PGI2-044 | 39192..39677 [+], 486 | hypothetical protein | |
| 46 | IS4 tnpA | 39874..40320 [-], 447 | transposase of IS4 | |
| 47 | tnp | 40395..40964 [-], 570 | transposase | |
| 48 | sul2 | 41054..41869 [+], 816 | dihydropteroate synthase type II | AR |
| 49 | glmM | 41956..42258 [+], 303 | phosphoglucosamine mutase GlmM | |
| 50 | rcr2 | 42434..43357 [-], 924 | rolling circle replicase Rcr2 | |
| 51 | lysR | 44039..44344 [-], 306 | transcriptional regulator | |
| 52 | floR | 44372..45586 [-], 1215 | florfenicol/chloramphenicol export protein FloR | AR |
| 53 | PGI2-052 | 45803..46687 [-], 885 | hypothetical protein | |
| 54 | PGI2-053 | 46718..47554 [-], 837 | hypothetical protein | |
| 55 | IS1006 tnpA | 47612..48316 [+], 705 | transposase | |
| 56 | Tn21 tnpA | 48811..51762 [-], 2952 | transposase of Tn21 | |
| 57 | tnpR | 51765..52325 [-], 561 | resolvase of Tn21 | |
| 58 | tnpM | 52451..52651 [-], 201 | TnpM | |
| 59 | intI1-2 | 53004..54017 [-], 1014 | IntI1 | Integrase |
| 60 | aadA2_2 | 54163..54954 [+], 792 | AadA2 | AR |
| 61 | lnuF | 55086..55907 [+], 822 | lincosamide nucleotidyltransferase | |
| 62 | qacEdelta1_2 | 56055..56402 [+], 348 | QacEdelta1 | |
| 63 | sul1_2 | 56396..57235 [+], 840 | Sul1 | AR |
| 64 | - | 57363..57863 [+], 501 | putative acetyltransferase | |
| 65 | - | 57887..58174 [+], 288 | hypothetical protein | |
| 66 | IS6100 tnpA | 58340..59134 [+], 795 | transposase of IS6100 | |
| 67 | PGI2-066 | 59352..59933 [+], 582 | hypothetical protein | |
| 68 | PGI2-067 | 60194..61066 [-], 873 | hypothetical protein | |
| 69 | PGI2-068 | 61056..62582 [-], 1527 | hypothetical protein | |
| 70 | PMI3124 | 63404..63868 [+], 465 | hypothetical protein | |

