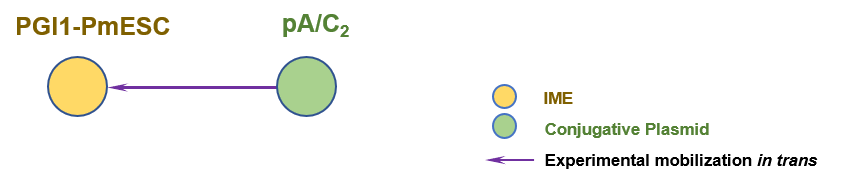
The graph information of PGI1-PmESC components from KU499917 |
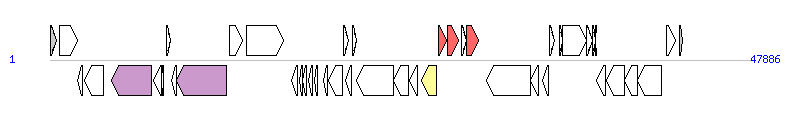 |
| Complete gene list of PGI1-PmESC from KU499917 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | trmE | 1..468 [+], 468 | tRNA modification GTPase | |
| 2 | int | 671..1885 [+], 1215 | phage integrase family protein | |
| 3 | - | 1882..2250 [-], 369 | hypothetical protein | |
| 4 | rep | 2385..3626 [-], 1242 | replication protein A | |
| 5 | - | 4241..6976 [-], 2736 | mating pair stabilization protein | TraN_F, T4SS component |
| 6 | - | 7063..7605 [-], 543 | putative regulator protein | |
| 7 | - | 7608..7796 [-], 189 | hypothetical protein | |
| 8 | - | 7968..8228 [+], 261 | hypothetical protein | |
| 9 | - | 8279..8635 [-], 357 | hypothetical protein | |
| 10 | - | 8632..12069 [-], 3438 | sex pilus assembly protein | TraG_F, T4SS component |
| 11 | - | 12278..13144 [+], 867 | hypothetical protein | |
| 12 | - | 13409..15994 [+], 2586 | helicase c2 | |
| 13 | - | 16500..16937 [-], 438 | hypothetical protein | |
| 14 | - | 17052..17243 [-], 192 | hypothetical protein | |
| 15 | - | 17303..17572 [-], 270 | hypothetical protein | |
| 16 | - | 17674..17991 [-], 318 | hypothetical protein | |
| 17 | - | 18070..18324 [-], 255 | hypothetical protein | |
| 18 | - | 18712..19008 [-], 297 | hypothetical protein | |
| 19 | - | 19028..19993 [-], 966 | phage integrase family protein | |
| 20 | - | 20083..20454 [+], 372 | hypothetical protein | |
| 21 | - | 20182..20604 [-], 423 | hypothetical protein | |
| 22 | - | 20680..20949 [+], 270 | hypothetical protein | |
| 23 | - | 20939..23482 [-], 2544 | Y4bN protein | |
| 24 | - | 23503..24492 [-], 990 | AAA ATPase | |
| 25 | res | 24571..25155 [-], 585 | resolvase | |
| 26 | intI1 | 25428..26441 [-], 1014 | IntI1 integrase | Integrase |
| 27 | aadB | 26588..27121 [+], 534 | aminoglycoside adenyltransferase | AR |
| 28 | aadA2 | 27179..27970 [+], 792 | aminoglycoside adenyltransferase | AR |
| 29 | qacEdelta1 | 28134..28481 [+], 348 | QacEdelta1 | |
| 30 | sul1 | 28475..29314 [+], 840 | dihydropteroate synthase | AR |
| 31 | tnpA | 29889..32855 [-], 2967 | transposase | |
| 32 | tnpR | 32859..33419 [-], 561 | resolvase | |
| 33 | merR | 33685..34119 [-], 435 | MerR regulatory protein (repressor/inducer) | |
| 34 | merT | 34191..34541 [+], 351 | mercury ion transport protein | |
| 35 | merF | 34824..35069 [+], 246 | mercuric ion transport protein | |
| 36 | merA | 35066..36712 [+], 1647 | mercuric ion reductase | |
| 37 | merD | 36729..37094 [+], 366 | modulator protein | |
| 38 | merE | 37091..37327 [+], 237 | MerE | |
| 39 | urf-2 | 37324..37341 [+], 18 | Urf-2 | |
| 40 | tniR | 37380..37994 [-], 615 | resolvase | |
| 41 | tniQ | 38055..39272 [-], 1218 | hypothetical protein | |
| 42 | tniB | 39269..40177 [-], 909 | putative ATP-binding protein | |
| 43 | tniA | 40180..41862 [-], 1683 | transposase | |
| 44 | - | 42148..42777 [+], 630 | hypothetical protein | |
| 45 | hipB | 43053..43245 [+], 193 | transcriptional regulator | |