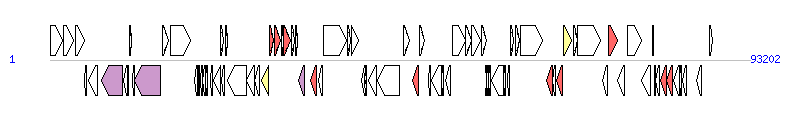The graph information of PGI1-PmCHA components from KJ411925 |
 |
| Complete gene list of PGI1-PmCHA from KJ411925 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | oxaA | 1..1641 [+], 1641 | inner membrane protein | |
| 2 | trmE | 1779..3143 [+], 1365 | tRNA modification GTPase | |
| 3 | int | 3346..4560 [+], 1215 | phage integrase family protein | |
| 4 | P002 | 4557..4925 [-], 369 | P002 | |
| 5 | rep | 5060..6301 [-], 1242 | replication protein A | |
| 6 | P004 | 6916..9651 [-], 2736 | mating pair stabilization protein | TraN_F, T4SS component |
| 7 | P005 | 9738..10280 [-], 543 | putative regulator protein | |
| 8 | P006 | 10283..10471 [-], 189 | hypothetical protein | |
| 9 | P007 | 10643..10903 [+], 261 | P007 | |
| 10 | P008 | 10954..11310 [-], 357 | P008 | |
| 11 | P009 | 11307..14744 [-], 3438 | sex pilus assembly protein | TraG_F, T4SS component |
| 12 | P010 | 14953..15819 [+], 867 | hypothetical protein | |
| 13 | P011 | 16084..18669 [+], 2586 | helicase c2 | |
| 14 | P012 | 19175..19612 [-], 438 | hypothetical protein | |
| 15 | P013 | 19727..19918 [-], 192 | P013 | |
| 16 | P014 | 19978..20247 [-], 270 | hypothetical protein | |
| 17 | P015 | 20349..20666 [-], 318 | P015 | |
| 18 | P016 | 20745..20999 [-], 255 | P016 | |
| 19 | P017 | 21387..21683 [-], 297 | P017 | |
| 20 | P018 | 21703..22668 [-], 966 | phage integrase family protein | |
| 21 | P019 | 22758..23129 [+], 372 | P019 | |
| 22 | P020 | 22857..23279 [-], 423 | P020 | |
| 23 | P021 | 23355..23624 [+], 270 | P021 | |
| 24 | P022 | 23614..26157 [-], 2544 | Y4bN protein | |
| 25 | P023 | 26178..27167 [-], 990 | AAA ATPase | |
| 26 | res | 27246..27830 [-], 585 | resolvase | |
| 27 | intI1 | 28103..29116 [-], 1014 | integrase | Integrase |
| 28 | aadB | 29263..29796 [+], 534 | aminoglycoside adenyltransferase | AR |
| 29 | aadA2 | 29854..30645 [+], 792 | aminoglycoside adenyltransferase | AR |
| 30 | qacEdelta1 | 30809..31156 [+], 348 | QacEdelta1 | |
| 31 | sul1 | 31150..31989 [+], 840 | dihydropteroate synthase | AR |
| 32 | orf5 | 32117..32617 [+], 501 | acetyltransferase | |
| 33 | orf6 | 32641..32928 [+], 288 | hypothetical protein | |
| 34 | istB | 33152..33937 [-], 786 | IstB | TrbB, T4SS component |
| 35 | blaTEM-135 | 34644..35504 [-], 861 | BlaTEM-135 beta-lactamase | AR |
| 36 | tnpR | 35687..36244 [-], 558 | TnpR resolvase | |
| 37 | tnpA | 36408..39413 [+], 3006 | TnpA transposase | |
| 38 | merR | 39608..40042 [+], 435 | Tn21 MerR | |
| 39 | tnpA | 40121..41125 [+], 1005 | IS4321 transposase | |
| 40 | insB | 41476..41979 [-], 504 | InsB | |
| 41 | insA | 41898..42173 [-], 276 | InsA | |
| 42 | tnpA | 42470..43474 [-], 1005 | IS4321 transposase | |
| 43 | tnpA | 43553..46519 [-], 2967 | TnpA transposase | |
| 44 | tnpA | 47099..47803 [+], 705 | IS26 transposase | |
| 45 | aphA1b | 48228..49043 [-], 816 | aminoglycoside phosphotransferase | AR |
| 46 | tnpA | 49173..49877 [+], 705 | IS26 transposase | |
| 47 | chbB | 50385..50711 [-], 327 | N,N'-diacetylchitobiose-specific PTS system EIIB component | |
| 48 | hipA | 50812..52125 [-], 1314 | putative regulatory protein | |
| 49 | hipB | 52126..52383 [-], 258 | transcriptional regulator | |
| 50 | P025 | 52659..53288 [-], 630 | P025 | |
| 51 | tniA | 53574..55256 [+], 1683 | transposase | |
| 52 | tniB | 55259..56167 [+], 909 | putative ATP-binding protein | |
| 53 | tniQ | 56164..57381 [+], 1218 | hypothetical protein | |
| 54 | tniR | 57442..58056 [+], 615 | resolvase | |
| 55 | urf-2 | 58095..58112 [-], 18 | urf-2 | |
| 56 | merE | 58109..58345 [-], 237 | MerE | |
| 57 | merD | 58342..58707 [-], 366 | modulator protein | |
| 58 | merA | 58724..60370 [-], 1647 | mercuric ion reductase | |
| 59 | merF | 60367..60612 [-], 246 | mercuric ion transport protein | |
| 60 | merT | 60895..61245 [-], 351 | mercury ion transport protein | |
| 61 | merR | 61317..61751 [+], 435 | MerR regulatory protein | |
| 62 | tnpR | 62017..62577 [+], 561 | resolvase | |
| 63 | tnpA | 62581..65547 [+], 2967 | transposase | |
| 64 | sul1 | 66122..66961 [-], 840 | dihydropteroate synthase | AR |
| 65 | qacEdelta1 | 66955..67302 [-], 348 | QacEdelta1 | |
| 66 | aadA1 | 67466..68257 [-], 792 | aminoglycoside adenyltransferase | AR |
| 67 | intI1 | 68403..69416 [+], 1014 | integrase | Integrase |
| 68 | tnpR | 69722..70279 [+], 558 | resolvase | |
| 69 | tnpA | 70282..73254 [+], 2973 | hybrid TnpA Tn1696/Tn1721 | |
| 70 | tetR | 73586..74236 [-], 651 | tetracycline resistance repressor | |
| 71 | tetA | 74342..75541 [+], 1200 | tetracycline resistance protein | AR |
| 72 | pecM | 75573..76457 [-], 885 | transporter protein | |
| 73 | tnpA' | 76946..78742 [+], 1797 | TnpA' transposase | |
| 74 | - | 78739..79956 [-], 1218 | DNA topoisomerase | |
| 75 | - | 80192..80338 [+], 147 | hypothetical protein | |
| 76 | - | 80472..80621 [-], 150 | hypothetical protein | |
| 77 | - | 80636..81106 [-], 471 | ParB protein | |
| 78 | strB | 81234..82070 [-], 837 | streptomycin phosphotransferase | AR |
| 79 | strA | 82070..82873 [-], 804 | streptomycin phosphotransferase | AR |
| 80 | orfB | 82980..83837 [-], 858 | ORF B | |
| 81 | orfD | 83834..84109 [-], 276 | ORF D | |
| 82 | tnpR | 84174..84719 [-], 546 | resolvase | |
| 83 | tnpA | 86024..86728 [-], 705 | IS26 transposase | |
| 84 | chbA | 87855..88202 [+], 348 | N,N'-diacetylchitobiose-specific PTS system EIIA component | |