
| 184_IME | |
| MTnPi2 | |
| - | |
| Prevotella intermedia s OMA14 | |
| 16557 bp | |
| TTGC NNNNN AA | |
| - | |
| - | |
| AP014597; AP014598 (complete IME sequence in this genome) | |
| - | |
| 767242..783798 | |
| - | |
| coordinates: 767485..768546; PIOMA14_I_0701 Family: MOBP |
The graph information of MTnPi2 components from AP014597 | |||||
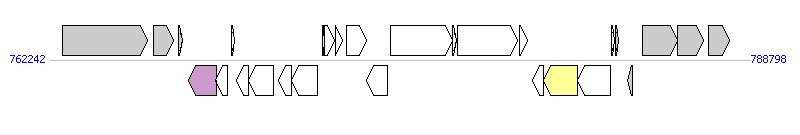 | |||||
| Complete gene list of MTnPi2 from AP014597 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation | |
| 1 | PIOMA14_I_0698 | 762705..765929 [+], 3225 | carbamoyl-phosphate synthase large subunit | ||
| 2 | PIOMA14_I_0699 | 766171..766929 [+], 759 | conserved hypothetical protein | ||
| 3 | PIOMA14_I_0700 | 767118..767264 [+], 147 | hypothetical protein | ||
| 4 | PIOMA14_I_0701 | 767485..768546 [-], 1062 | relaxase/mobilization nuclease | Relaxase | |
| 5 | PIOMA14_I_0702 | 768530..768961 [-], 432 | conserved hypothetical protein | ||
| 6 | PIOMA14_I_0703 | 769116..769229 [+], 114 | hypothetical protein | ||
| 7 | PIOMA14_I_0704 | 769323..769784 [-], 462 | conserved hypothetical protein with DUF3408 domain | ||
| 8 | PIOMA14_I_0705 | 769789..770739 [-], 951 | probable DNA primase | ||
| 9 | PIOMA14_I_0706 | 770922..771401 [-], 480 | partial virulence-associated E family protein | ||
| 10 | PIOMA14_I_0707 | 771388..772374 [-], 987 | partial virulence-associated E family protein | ||
| 11 | PIOMA14_I_0708 | 772575..772655 [+], 81 | hypothetical protein | ||
| 12 | PIOMA14_I_0709 | 772670..773032 [+], 363 | hypothetical protein with HTH17 domain | ||
| 13 | PIOMA14_I_0710 | 773063..773353 [+], 291 | hypothetical protein | ||
| 14 | PIOMA14_I_0711 | 773481..774230 [+], 750 | conserved hypothetical protein with ThiF domain | ||
| 15 | PIOMA14_I_0712 | 774245..775051 [-], 807 | putative LuxR family transcriptional regulator | ||
| 16 | PIOMA14_I_0713 | 775143..777494 [+], 2352 | conserved hypothetical protein with Can B domain | ||
| 17 | PIOMA14_I_0714 | 777495..777680 [+], 186 | hypothetical protein | ||
| 18 | PIOMA14_I_0715 | 777698..779992 [+], 2295 | conserved hypothetical protein with Can B domain | ||
| 19 | PIOMA14_I_0716 | 780066..780374 [+], 309 | hypothetical protein | ||
| 20 | PIOMA14_I_0717 | 780548..780964 [-], 417 | conserved hypothetical protein | ||
| 21 | PIOMA14_I_0718 | 780961..782256 [-], 1296 | integrase | Integrase | |
| 22 | PIOMA14_I_0719 | 782269..783498 [-], 1230 | integrase | ||
| 23 | PIOMA14_I_0720 | 783550..783645 [+], 96 | conserved hypothetical protein | ||
| 24 | PIOMA14_I_0721 | 783711..783806 [+], 96 | hypothetical protein | ||
| 25 | PIOMA14_I_0722 | 784132..784323 [-], 192 | hypothetical protein | ||
| 26 | PIOMA14_I_0723 | 784714..786054 [+], 1341 | conserved hypothetical protein with UPF004 and RadicalSAM domain | ||
| 27 | PIOMA14_I_0724 | 786051..787052 [+], 1002 | glycosyltransferase family 2 | ||
| 28 | PIOMA14_I_0725 | 787209..788015 [+], 807 | conserved hypothetical protein | ||
The graph information of MTnPi2 components from AP014597 | |||||
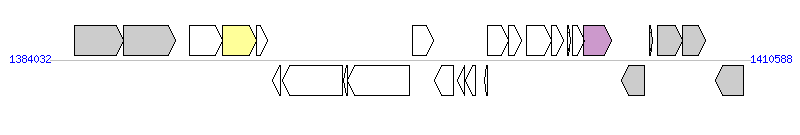 | |||||
| Complete gene list of MTnPi2 from AP014597 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation | |
| 1 | PIOMA14_I_1254 | 1384954..1386813 [+], 1860 | ABC transporter ATP-binding protein | ||
| 2 | PIOMA14_I_1255 | 1386818..1388791 [+], 1974 | sulfatase | ||
| 3 | PIOMA14_I_1256 | 1389332..1390561 [+], 1230 | integrase | ||
| 4 | PIOMA14_I_1257 | 1390574..1391869 [+], 1296 | integrase | Integrase | |
| 5 | PIOMA14_I_1258 | 1391866..1392282 [+], 417 | conserved hypothetical protein | ||
| 6 | PIOMA14_I_1259 | 1392456..1392764 [-], 309 | hypothetical protein | ||
| 7 | PIOMA14_I_1260 | 1392838..1395132 [-], 2295 | conserved hypothetical protein with Can-B2 domain | ||
| 8 | PIOMA14_I_1261 | 1395150..1395335 [-], 186 | hypothetical protein | ||
| 9 | PIOMA14_I_1262 | 1395336..1397687 [-], 2352 | conserved hypothetical protein with Can-B2 domain | ||
| 10 | PIOMA14_I_1263 | 1397779..1398585 [+], 807 | putative LuxR family transcriptional regulator | ||
| 11 | PIOMA14_I_1264 | 1398600..1399349 [-], 750 | conserved hypothetical protein with ThiF domain | ||
| 12 | PIOMA14_I_1265 | 1399477..1399767 [-], 291 | hypothetical protein | ||
| 13 | PIOMA14_I_1266 | 1399798..1400160 [-], 363 | hypothetical protein with HTH17 domain | ||
| 14 | PIOMA14_I_1267 | 1400498..1400638 [-], 141 | hypothetical protein | ||
| 15 | PIOMA14_I_1268 | 1400642..1401442 [+], 801 | partial virulence-associated E family protein | ||
| 16 | PIOMA14_I_1269 | 1401429..1401908 [+], 480 | partial virulence-associated E family protein | ||
| 17 | PIOMA14_I_1270 | 1402091..1403041 [+], 951 | probable DNA primase | ||
| 18 | PIOMA14_I_1271 | 1403046..1403507 [+], 462 | conserved hypothetical protein with DUF3408 domain | ||
| 19 | PIOMA14_I_1272 | 1403647..1403787 [+], 141 | hypothetical protein | ||
| 20 | PIOMA14_I_1273 | 1403869..1404300 [+], 432 | conserved hypothetical protein | ||
| 21 | PIOMA14_I_1274 | 1404284..1405345 [+], 1062 | relaxase/mobilisation nuclease | Relaxase | |
| 22 | PIOMA14_I_1275 | 1405714..1406601 [-], 888 | conserved hypothetical protein with EamA domain | ||
| 23 | PIOMA14_I_1276 | 1406776..1406895 [+], 120 | hypothetical protein | ||
| 24 | PIOMA14_I_1277 | 1407065..1408012 [+], 948 | putative lipid A biosynthesis acyltransferase | ||
| 25 | PIOMA14_I_1278 | 1408035..1408898 [+], 864 | conserved hypothetical protein with LicD domain | ||
| 26 | PIOMA14_I_1279 | 1409283..1410332 [-], 1050 | conserved hypothetical protein with Glyphos-transf domain | ||
The graph information of MTnPi2 components from AP014598 | |||||
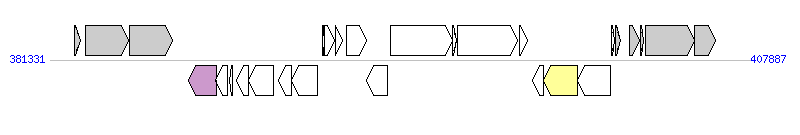 | |||||
| Complete gene list of MTnPi2 from AP014598 | |||||
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation | |
| 1 | PIOMA14_II_0320 | 382269..382475 [+], 207 | hypothetical protein | ||
| 2 | PIOMA14_II_0321 | 382675..384318 [+], 1644 | peptidase M23 family | ||
| 3 | PIOMA14_II_0322 | 384353..385990 [+], 1638 | peptidase C69 family/dipeptidase | ||
| 4 | PIOMA14_II_0323 | 386574..387635 [-], 1062 | relaxase/mobilization nuclease | Relaxase | |
| 5 | PIOMA14_II_0324 | 387619..388050 [-], 432 | conserved hypothetical protein | ||
| 6 | PIOMA14_II_0325 | 388132..388272 [-], 141 | hypothetical protein | ||
| 7 | PIOMA14_II_0326 | 388412..388873 [-], 462 | conserved hypothetical protein with DUF3408 domain | ||
| 8 | PIOMA14_II_0327 | 388878..389828 [-], 951 | probable DNA primase | ||
| 9 | PIOMA14_II_0328 | 390011..390490 [-], 480 | partial virulence-associated E family protein | ||
| 10 | PIOMA14_II_0329 | 390477..391463 [-], 987 | partial virulence-associated E family protein | ||
| 11 | PIOMA14_II_0330 | 391664..391744 [+], 81 | hypothetical protein | ||
| 12 | PIOMA14_II_0331 | 391759..392121 [+], 363 | hypothetical protein with HTH17 domain | ||
| 13 | PIOMA14_II_0332 | 392152..392442 [+], 291 | hypothetical protein | ||
| 14 | PIOMA14_II_0333 | 392570..393319 [+], 750 | conserved hypothetical protein with ThiF domain | ||
| 15 | PIOMA14_II_0334 | 393334..394140 [-], 807 | putative transcriptional regulator LuxR family | ||
| 16 | PIOMA14_II_0335 | 394232..396583 [+], 2352 | conserved hypothetical protein with CnaB2 domain | ||
| 17 | PIOMA14_II_0336 | 396584..396769 [+], 186 | hypothetical protein | ||
| 18 | PIOMA14_II_0337 | 396787..399081 [+], 2295 | conserved hypothetical protein with CnaB2 domain | ||
| 19 | PIOMA14_II_0338 | 399155..399463 [+], 309 | hypothetical protein | ||
| 20 | PIOMA14_II_0339 | 399637..400053 [-], 417 | conserved hypothetical protein | ||
| 21 | PIOMA14_II_0340 | 400050..401345 [-], 1296 | integrase | Integrase | |
| 22 | PIOMA14_II_0341 | 401358..402587 [-], 1230 | integrase | ||
| 23 | PIOMA14_II_0342 | 402639..402734 [+], 96 | conserved hypothetical protein | ||
| 24 | PIOMA14_II_0343 | 402800..402970 [+], 171 | hypothetical protein | ||
| 25 | PIOMA14_II_0344 | 403309..403707 [+], 399 | hypothetical protein | ||
| 26 | PIOMA14_II_0345 | 403740..403835 [+], 96 | conserved hypothetical protein | ||
| 27 | PIOMA14_II_0346 | 403926..405776 [+], 1851 | Rhs element Vgr protein | ||
| 28 | PIOMA14_II_0347 | 405780..406571 [+], 792 | hypothetical protein | ||
| (1) Guedon G; Libante V; Coluzzi C; Payot S; Leblond-Bourget N (2017). The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes (Basel). 8(11). [PudMed:29165361] |
|
(2) Naito M; Ogura Y; Itoh T; Shoji M; Okamoto M; Hayashi T; Nakayama K (2016). The complete genome sequencing of Prevotella intermedia strain OMA14 and a subsequent fine-scale, intra-species genomic comparison reveal an unusual amplification of conjugative and mobile transposons and identify a novel Prevotella-lineage-specific repeat. DNA Res. 23(1):11-9. [PudMed:26645327] |