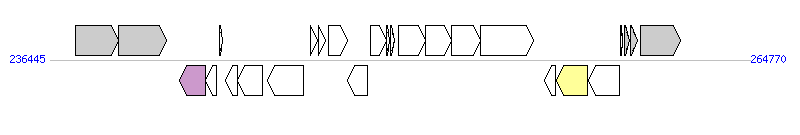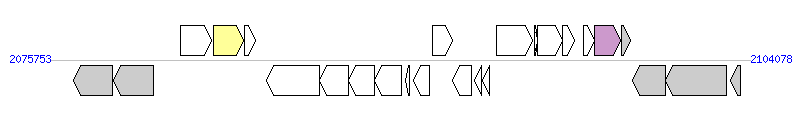The graph information of MTnPi1 components from AP014597 |
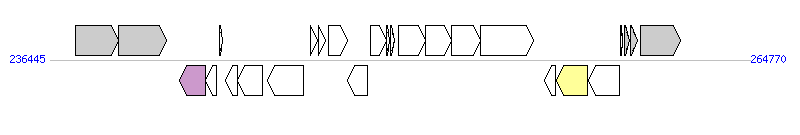 |
| Complete gene list of MTnPi1 from AP014597 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | PIOMA14_I_0215 | 237479..239203 [+], 1725 | single-stranded-DNA-specific exonuclease RecJ | |
| 2 | PIOMA14_I_0216 | 239236..241152 [+], 1917 | ATP-dependent helicase RecQ | |
| 3 | PIOMA14_I_0217 | 241690..242751 [-], 1062 | relaxase/mobilization nuclease | Relaxase |
| 4 | PIOMA14_I_0218 | 242735..243166 [-], 432 | conserved hypothetical protein | |
| 5 | PIOMA14_I_0219 | 243321..243425 [+], 105 | hypothetical protein | |
| 6 | PIOMA14_I_0220 | 243549..244019 [-], 471 | conserved hypothetical protein with DUF3408 domain | |
| 7 | PIOMA14_I_0221 | 244022..245026 [-], 1005 | probable DNA primase | |
| 8 | PIOMA14_I_0222 | 245263..246693 [-], 1431 | virulence-associated protein E | |
| 9 | PIOMA14_I_0223 | 246985..247269 [+], 285 | hypothetical protein with helix-turn-helix domain | |
| 10 | PIOMA14_I_0224 | 247300..247590 [+], 291 | hypothetical protein with helix-turn-helix domain | |
| 11 | PIOMA14_I_0225 | 247718..248467 [+], 750 | conserved hypothetical protein with ThiF family domain | |
| 12 | PIOMA14_I_0226 | 248482..249288 [-], 807 | LuxR family transcriptional regulator | |
| 13 | PIOMA14_I_0227 | 249428..250075 [+], 648 | conserved hypothetical protein | |
| 14 | PIOMA14_I_0228 | 250056..250187 [+], 132 | hypothetical protein | |
| 15 | PIOMA14_I_0229 | 250220..250372 [+], 153 | hypothetical protein | |
| 16 | PIOMA14_I_0230 | 250543..251643 [+], 1101 | conserved hypothetical protein | |
| 17 | PIOMA14_I_0231 | 251621..252685 [+], 1065 | conserved hypothetical protein | |
| 18 | PIOMA14_I_0232 | 252682..253875 [+], 1194 | probable arginase/agmatinase | |
| 19 | PIOMA14_I_0233 | 253872..255995 [+], 2124 | conserved hypothetical protein | |
| 20 | PIOMA14_I_0234 | 256469..256885 [-], 417 | hypothetical protein | |
| 21 | PIOMA14_I_0235 | 256925..258175 [-], 1251 | integrase | Integrase |
| 22 | PIOMA14_I_0236 | 258241..259470 [-], 1230 | integrase | |
| 23 | PIOMA14_I_0237 | 259522..259617 [+], 96 | conserved hypothetical protein | |
| 24 | PIOMA14_I_0238 | 259683..259886 [+], 204 | hypothetical protein | |
| 25 | groES | 259953..260222 [+], 270 | chaperonin GroES | |
| 26 | groEL | 260345..261970 [+], 1626 | chaperonin GroEL | |
The graph information of MTnPi1 components from AP014597 |
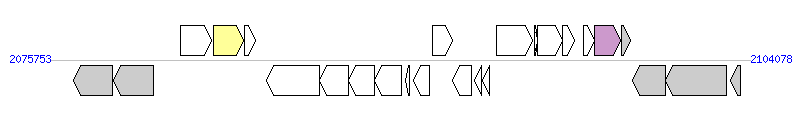 |
| Complete gene list of MTnPi1 from AP014597 |
| # | Gene | Coordinates [+/-], size (bp) | Product | *Reannotation  |
| 1 | PIOMA14_I_1839 | 2076695..2078299 [-], 1605 | conserved hypothetical protein with BACON domain | |
| 2 | PIOMA14_I_1840 | 2078335..2079954 [-], 1620 | conserved hypothetical protein with BACON domain | |
| 3 | PIOMA14_I_1841 | 2081053..2082282 [+], 1230 | integrase | |
| 4 | PIOMA14_I_1842 | 2082348..2083598 [+], 1251 | integrase | Integrase |
| 5 | PIOMA14_I_1843 | 2083638..2084054 [+], 417 | hypothetical protein | |
| 6 | PIOMA14_I_1844 | 2084528..2086651 [-], 2124 | conserved hypothetical protein | |
| 7 | PIOMA14_I_1845 | 2086648..2087841 [-], 1194 | probable arginase/agmatinase | |
| 8 | PIOMA14_I_1846 | 2087838..2088902 [-], 1065 | conserved hypothetical protein | |
| 9 | PIOMA14_I_1847 | 2088880..2089980 [-], 1101 | conserved hypothetical protein | |
| 10 | PIOMA14_I_1848 | 2090151..2090303 [-], 153 | hypothetical protein | |
| 11 | PIOMA14_I_1849 | 2090448..2091095 [-], 648 | conserved hypothetical protein | |
| 12 | PIOMA14_I_1850 | 2091235..2092041 [+], 807 | LuxR family transcriptional regulator | |
| 13 | PIOMA14_I_1851 | 2092056..2092805 [-], 750 | conserved hypothetical protein with ThiF domain | |
| 14 | PIOMA14_I_1852 | 2092933..2093223 [-], 291 | hypothetical protein with helix-turn-helix domain | |
| 15 | PIOMA14_I_1853 | 2093254..2093538 [-], 285 | hypothetical protein with helix-turn-helix domain | |
| 16 | PIOMA14_I_1854 | 2093830..2095260 [+], 1431 | virulence-associated E family protein | |
| 17 | PIOMA14_I_1855 | 2095357..2095485 [+], 129 | hypothetical protein | |
| 18 | PIOMA14_I_1856 | 2095497..2096501 [+], 1005 | probable DNA primase | |
| 19 | PIOMA14_I_1857 | 2096504..2096974 [+], 471 | conserved hypothetical protein with DF3408 domain | |
| 20 | PIOMA14_I_1858 | 2097357..2097788 [+], 432 | conserved hypothetical protein | |
| 21 | PIOMA14_I_1859 | 2097772..2098833 [+], 1062 | relaxase/mobilization nuclease | Relaxase |
| 22 | PIOMA14_I_1860 | 2098867..2099226 [+], 360 | hypothetical protein | |
| 23 | PIOMA14_I_1861 | 2099341..2100639 [-], 1299 | putative DNA recombination protein RmuC | |
| 24 | PIOMA14_I_1862 | 2100651..2103131 [-], 2481 | alanine racemase/Mur ligase domain protein | |
| 25 | PIOMA14_I_1863 | 2103272..2103700 [-], 429 | conserved hypothetical protein | |